Mash Missing Sample Size
Yuxin Zou
2018-3-1
Last updated: 2018-03-06
Code version: 193208d
The weights from data R = 50, N = 400 are non-zero. Increasing the sample size to 800 and 3200, the weights shrink to 0. Decreasing the sampel size to 400 provides the correct weights as well.
N = 400
library(mashr)Loading required package: ashrset.seed(2018)
data = simple_sims(100, 50, err_sd = 0.1)
mash.data = mash_set_data(data$Bhat, data$Shat)
U.c = cov_canonical(mash.data)
mash.model = mash(mash.data, U.c) - Computing 400 x 1101 likelihood matrix.
- Likelihood calculations took 1.00 seconds.
- Fitting model with 1101 mixture components.Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtolWarning in mixIP(matrix_lik = structure(c(0.0182689419778344,
0.023990286246669, : Optimization step yields mixture weights that are
either too small, or negative; weights have been corrected and renormalized
after the optimization. - Model fitting took 1.94 seconds.
- Computing posterior matrices.
- Computation allocated took 1.27 seconds.barplot(get_estimated_pi(mash.model), las=2, cex.names = 0.7)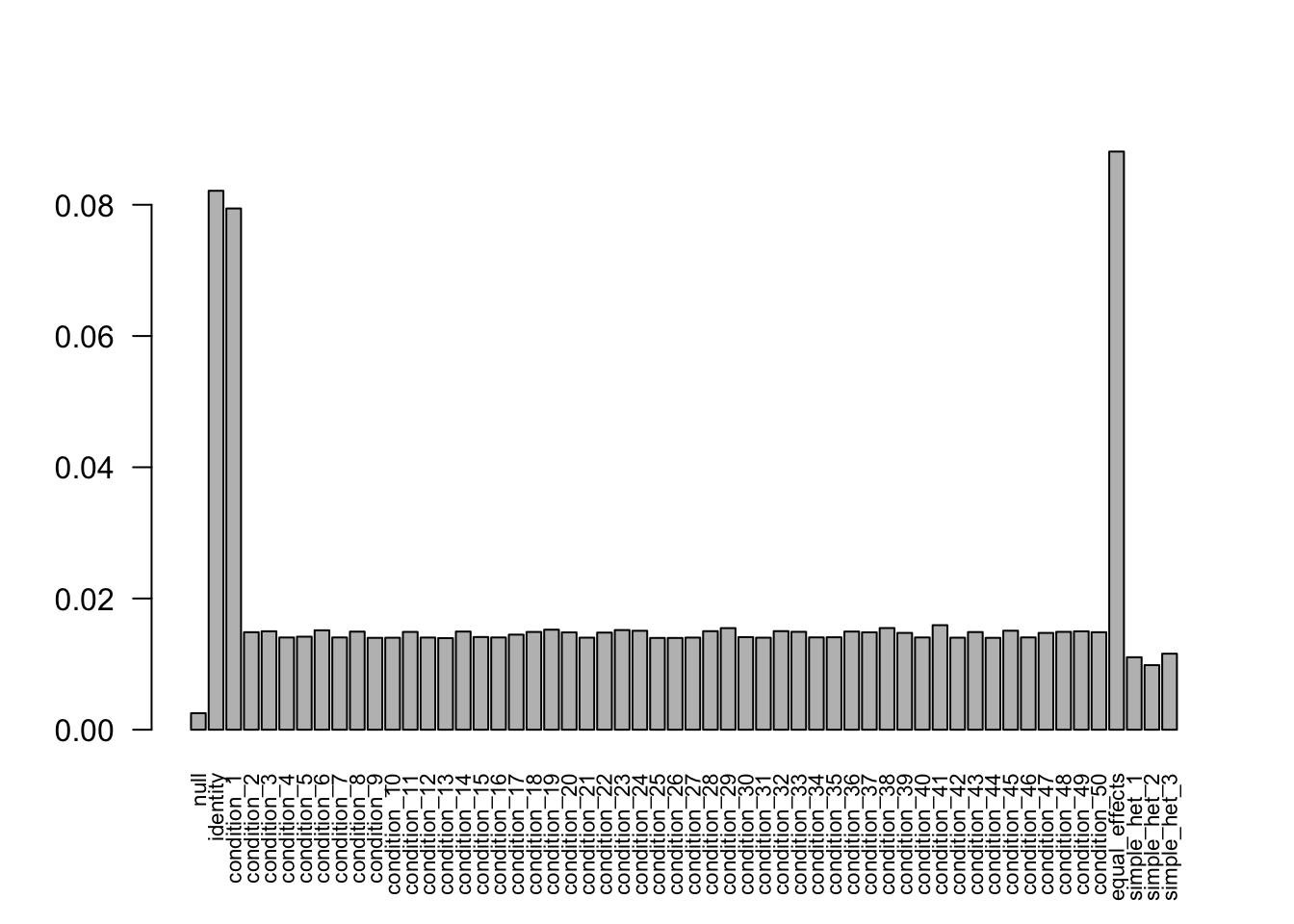
Increase to N = 800
set.seed(2018)
data = simple_sims(200, 50, err_sd = 0.1)
mash.data = mash_set_data(data$Bhat, data$Shat)
U.c = cov_canonical(mash.data)
mash.model = mash(mash.data, U.c) - Computing 800 x 1156 likelihood matrix.
- Likelihood calculations took 1.73 seconds.
- Fitting model with 1156 mixture components.Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtolWarning in mixIP(matrix_lik = structure(c(0.0183236802594066,
0.0842224653599791, : Optimization step yields mixture weights that are
either too small, or negative; weights have been corrected and renormalized
after the optimization. - Model fitting took 4.59 seconds.
- Computing posterior matrices.
- Computation allocated took 2.54 seconds.barplot(get_estimated_pi(mash.model), las=2, cex.names = 0.7)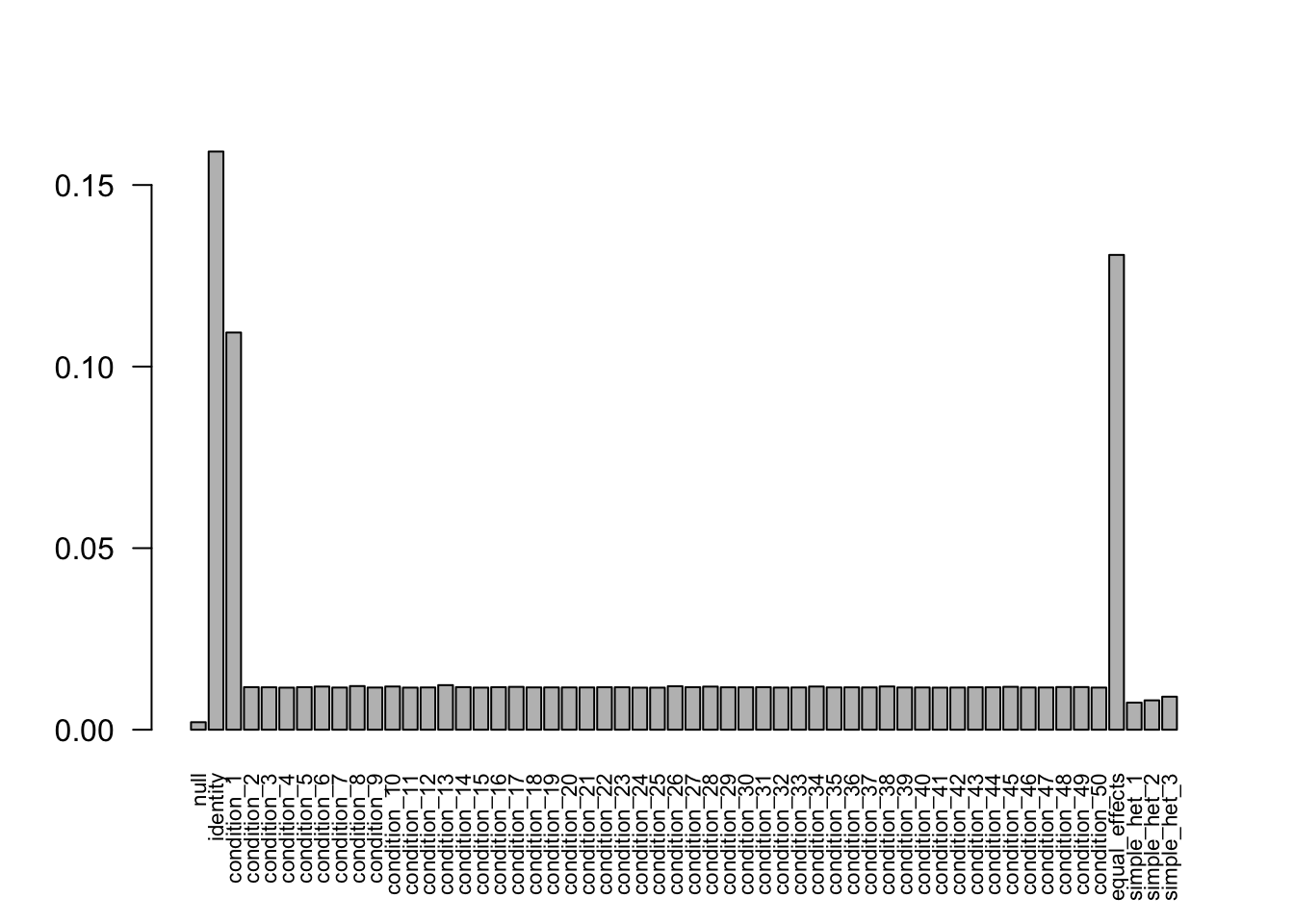
There are some shinkage for the small weights.
Increase to N = 3200
set.seed(2018)
data = simple_sims(800, 50, err_sd = 0.1)
mash.data = mash_set_data(data$Bhat, data$Shat)
U.c = cov_canonical(mash.data)
mash.model = mash(mash.data, U.c) - Computing 3200 x 1156 likelihood matrix.
- Likelihood calculations took 7.04 seconds.
- Fitting model with 1156 mixture components.Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtolWarning in mixIP(matrix_lik = structure(c(0.195406888247947,
0.198551348399632, : Optimization step yields mixture weights that are
either too small, or negative; weights have been corrected and renormalized
after the optimization. - Model fitting took 5.28 seconds.
- Computing posterior matrices.
- Computation allocated took 3.97 seconds.barplot(get_estimated_pi(mash.model), las=2, cex.names = 0.7)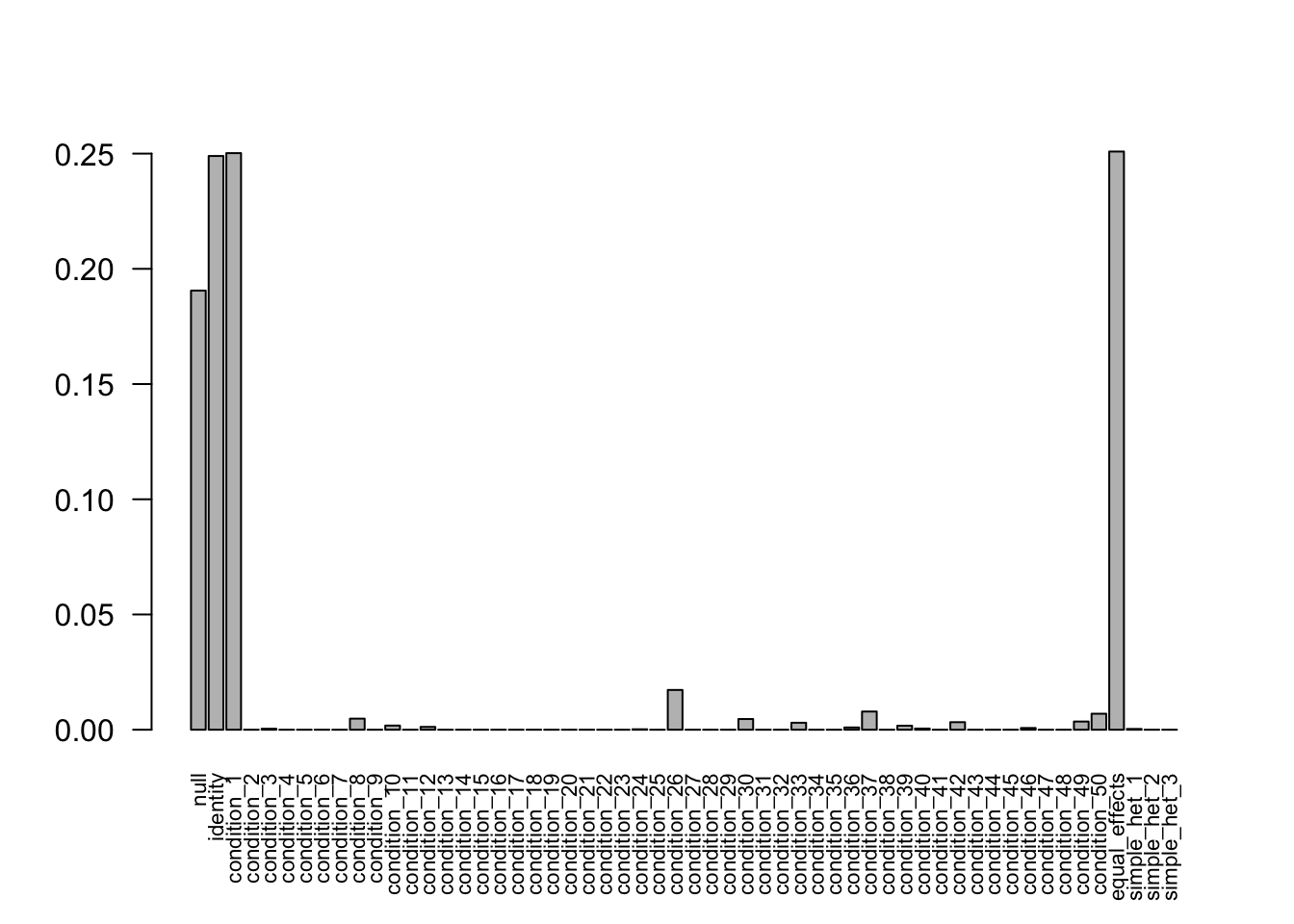
Decrease to N = 300
set.seed(2018)
data = simple_sims(75, 50, err_sd = 0.1)
mash.data = mash_set_data(data$Bhat, data$Shat)
U.c = cov_canonical(mash.data)
mash.model = mash(mash.data, U.c) - Computing 300 x 1101 likelihood matrix.
- Likelihood calculations took 0.62 seconds.
- Fitting model with 1101 mixture components.Warning in REBayes::KWDual(A, rep(1, k), normalize(w), control = control): estimated mixing distribution has some negative values:
consider reducing rtolWarning in mixIP(matrix_lik = structure(c(0.00730053524366138,
0.268626142814483, : Optimization step yields mixture weights that are
either too small, or negative; weights have been corrected and renormalized
after the optimization. - Model fitting took 1.11 seconds.
- Computing posterior matrices.
- Computation allocated took 0.10 seconds.barplot(get_estimated_pi(mash.model), las=2, cex.names = 0.7)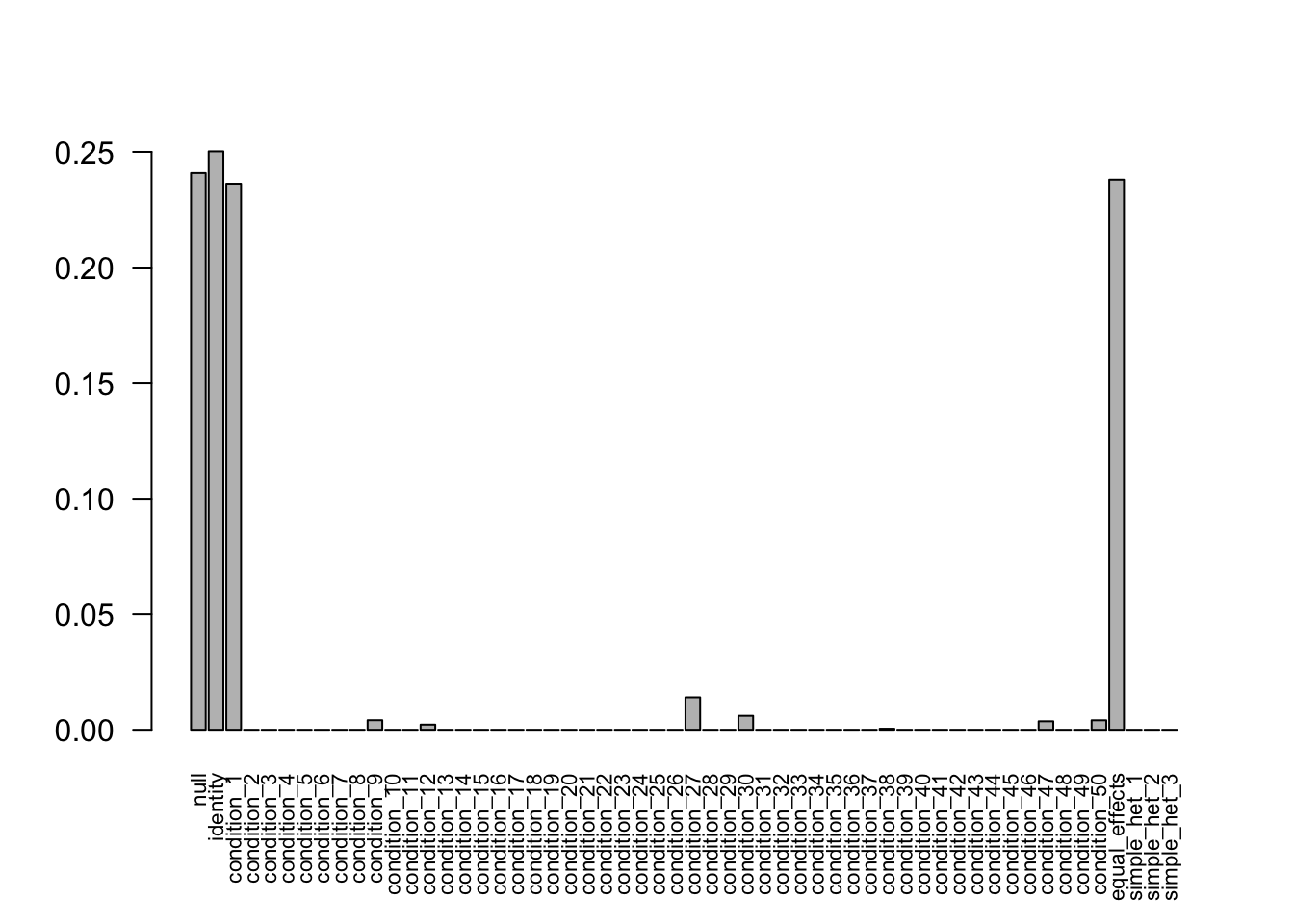
The weights learnd from N=300 model is correct.
Session information
sessionInfo()R version 3.4.3 (2017-11-30)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.3
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] mashr_0.2-6 ashr_2.2-7
loaded via a namespace (and not attached):
[1] Rcpp_0.12.15 knitr_1.20 magrittr_1.5
[4] REBayes_1.2 MASS_7.3-47 doParallel_1.0.11
[7] pscl_1.5.2 SQUAREM_2017.10-1 lattice_0.20-35
[10] foreach_1.4.4 plyr_1.8.4 stringr_1.3.0
[13] tools_3.4.3 parallel_3.4.3 grid_3.4.3
[16] rmeta_2.16 git2r_0.20.0 htmltools_0.3.6
[19] iterators_1.0.9 assertthat_0.2.0 yaml_2.1.17
[22] rprojroot_1.2 digest_0.6.13 Matrix_1.2-12
[25] codetools_0.2-15 evaluate_0.10.1 rmarkdown_1.8
[28] stringi_1.1.6 compiler_3.4.3 Rmosek_8.0.69
[31] backports_1.1.2 mvtnorm_1.0-7 truncnorm_1.0-8 This R Markdown site was created with workflowr