MASH Null
Yuxin Zou
2018-08-11
Last updated: 2018-08-21
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(1)The command
set.seed(1)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 3eb1f9b
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: analysis/.DS_Store Ignored: analysis/.Rhistory Ignored: analysis/include/.DS_Store Ignored: data/.DS_Store Ignored: docs/.DS_Store Ignored: output/.DS_Store Untracked files: Untracked: analysis/Classify.Rmd Untracked: analysis/EstimateCorMaxEM.Rmd Untracked: analysis/EstimateCorMaxEMGD.Rmd Untracked: analysis/EstimateCorPrior.Rmd Untracked: analysis/EstimateCorSol.Rmd Untracked: analysis/HierarchicalFlashSim.Rmd Untracked: analysis/Mash_GTEx.Rmd Untracked: analysis/MeanAsh.Rmd Untracked: analysis/OutlierDetection.Rmd Untracked: analysis/OutlierDetection2.Rmd Untracked: analysis/OutlierDetection3.Rmd Untracked: analysis/OutlierDetection4.Rmd Untracked: analysis/Test.Rmd Untracked: analysis/mash_missing_row.Rmd Untracked: code/MashClassify.R Untracked: code/MashCorResult.R Untracked: code/MashNULLCorResult.R Untracked: code/MashSource.R Untracked: code/Weight_plot.R Untracked: code/addemV.R Untracked: code/estimate_cor.R Untracked: code/generateDataV.R Untracked: code/johnprocess.R Untracked: code/sim_mean_sig.R Untracked: code/summary.R Untracked: data/Blischak_et_al_2015/ Untracked: data/scale_data.rds Untracked: docs/figure/Classify.Rmd/ Untracked: docs/figure/OutlierDetection.Rmd/ Untracked: docs/figure/OutlierDetection2.Rmd/ Untracked: docs/figure/OutlierDetection3.Rmd/ Untracked: docs/figure/Test.Rmd/ Untracked: docs/figure/mash_missing_whole_row_5.Rmd/ Untracked: docs/include/ Untracked: output/AddEMV/ Untracked: output/CovED_UKBio_strong.rds Untracked: output/CovED_UKBio_strong_Z.rds Untracked: output/Flash_UKBio_strong.rds Untracked: output/MASH.10.em2.result.rds Untracked: output/MASH.10.mle.result.rds Untracked: output/MASHNULL.V.result.1.rds Untracked: output/MASHNULL.V.result.10.rds Untracked: output/MASHNULL.V.result.11.rds Untracked: output/MASHNULL.V.result.12.rds Untracked: output/MASHNULL.V.result.13.rds Untracked: output/MASHNULL.V.result.14.rds Untracked: output/MASHNULL.V.result.15.rds Untracked: output/MASHNULL.V.result.16.rds Untracked: output/MASHNULL.V.result.17.rds Untracked: output/MASHNULL.V.result.18.rds Untracked: output/MASHNULL.V.result.19.rds Untracked: output/MASHNULL.V.result.2.rds Untracked: output/MASHNULL.V.result.20.rds Untracked: output/MASHNULL.V.result.3.rds Untracked: output/MASHNULL.V.result.4.rds Untracked: output/MASHNULL.V.result.5.rds Untracked: output/MASHNULL.V.result.6.rds Untracked: output/MASHNULL.V.result.7.rds Untracked: output/MASHNULL.V.result.8.rds Untracked: output/MASHNULL.V.result.9.rds Untracked: output/MashCorSim--midway/ Untracked: output/Mash_EE_Cov_0_plusR1.rds Untracked: output/UKBio_mash_model.rds Unstaged changes: Modified: analysis/Mash_UKBio.Rmd Modified: analysis/mash_missing_samplesize.Rmd Modified: output/Flash_T2_0.rds Modified: output/Flash_T2_0_mclust.rds Modified: output/Mash_model_0_plusR1.rds Modified: output/PresiAddVarCol.rds
Expand here to see past versions:
library(mashr)Loading required package: ashrsource('../code/estimate_cor.R')
source('../code/generateDataV.R')
library(knitr)
library(kableExtra)V identity
\[ V = I_{5} \]
set.seed(1)
n = 3000; p = 5
U0 = matrix(0, p,p)
Utrue = list(U0 = U0)
V = diag(p)
data = generate_data(n=n, p=p, V = V, Utrue = Utrue)mash model
samp = 1:(n/2)
if(is.null(dim(data$Shat))){
data$Shat = matrix(data$Shat, n, p)
}
data.samp = lapply(data, function(l) l[samp,])
m.data = mash_set_data(Bhat = data.samp$Bhat, Shat = data.samp$Shat)
U.c = cov_canonical(m.data)
# m.1by1 = mash_1by1(m.data)
# strong = get_significant_results(m.1by1)
Vhat = estimate_null_correlation(m.data)
Vhat.em = estimateV(m.data, U.c, init_rho = c(-0.5,0,0.5),tol=1e-4, optmethod='em2')$V
m.data.em = mash_set_data(Bhat=data.samp$Bhat, Shat = data.samp$Shat, V = Vhat.em)
m.model.em = mash(m.data.em, U.c, verbose=FALSE)res = rbind(c(norm(Vhat.em-diag(5), 'F'), get_loglik(m.model.em), length(get_significant_results(m.model.em))))
colnames(res) = c('F.error', 'loglik', '# significance')
row.names(res) = 'EM'
res %>% kable() %>% kable_styling()| F.error | loglik | # significance | |
|---|---|---|---|
| EM | 0.1178653 | -10767.46 | 0 |
V not idendity
We simulate 20 null data sets with non identity V, and check the mash results.
set.seed(1)
n = 2000; p = 5
U0 = matrix(0, p,p)
Utrue = list(U0 = U0)
for(t in 1:20){
V = clusterGeneration::rcorrmatrix(p)
data = generate_data(n=n, p=p, V = V, Utrue = Utrue)
samp = 1:(n/2)
if(is.null(dim(data$Shat))){
data$Shat = matrix(data$Shat, n, p)
}
data.samp = lapply(data, function(l) l[samp,])
m.data = mash_set_data(Bhat = data.samp$Bhat, Shat = data.samp$Shat)
U.c = cov_canonical(m.data)
Vhat = estimate_null_correlation(m.data, apply_lower_bound = FALSE)
Vhat.em = estimateV(m.data, U.c, init_rho = c(-0.5,0,0.5), tol=1e-4, optmethod='em2')
R <- tryCatch(chol(Vhat.em$V),error = function (e) FALSE)
if(!is.matrix(R)){
pd = FALSE
Vhat.em$V = as.matrix(Matrix::nearPD(Vhat.em$V, conv.norm.type = 'F', keepDiag = TRUE)$mat)
}else{
pd = TRUE
}
m.data.trunc = mash_set_data(data.samp$Bhat, data.samp$Shat, V = Vhat)
m.model.trunc = mash(m.data.trunc, U.c)
m.data.em = mash_set_data(Bhat=data.samp$Bhat, Shat = data.samp$Shat, V = Vhat.em$V)
m.model.em = mash(m.data.em, U.c)
saveRDS(list(pd = pd, V.true = V, V.em = Vhat.em, V.trunc = Vhat, data = data, sample = samp, model.trunc = m.model.trunc, model.em = m.model.em),
paste0('../output/MASHNULL.V.result.',t,'.rds'))
}files = dir("../output/"); files = files[grep("MASHNULL.V.result",files)]
times = length(files)
result = vector(mode="list",length = times)
for(i in 1:times) {
result[[i]] = readRDS(paste("../output/", files[[i]], sep=""))
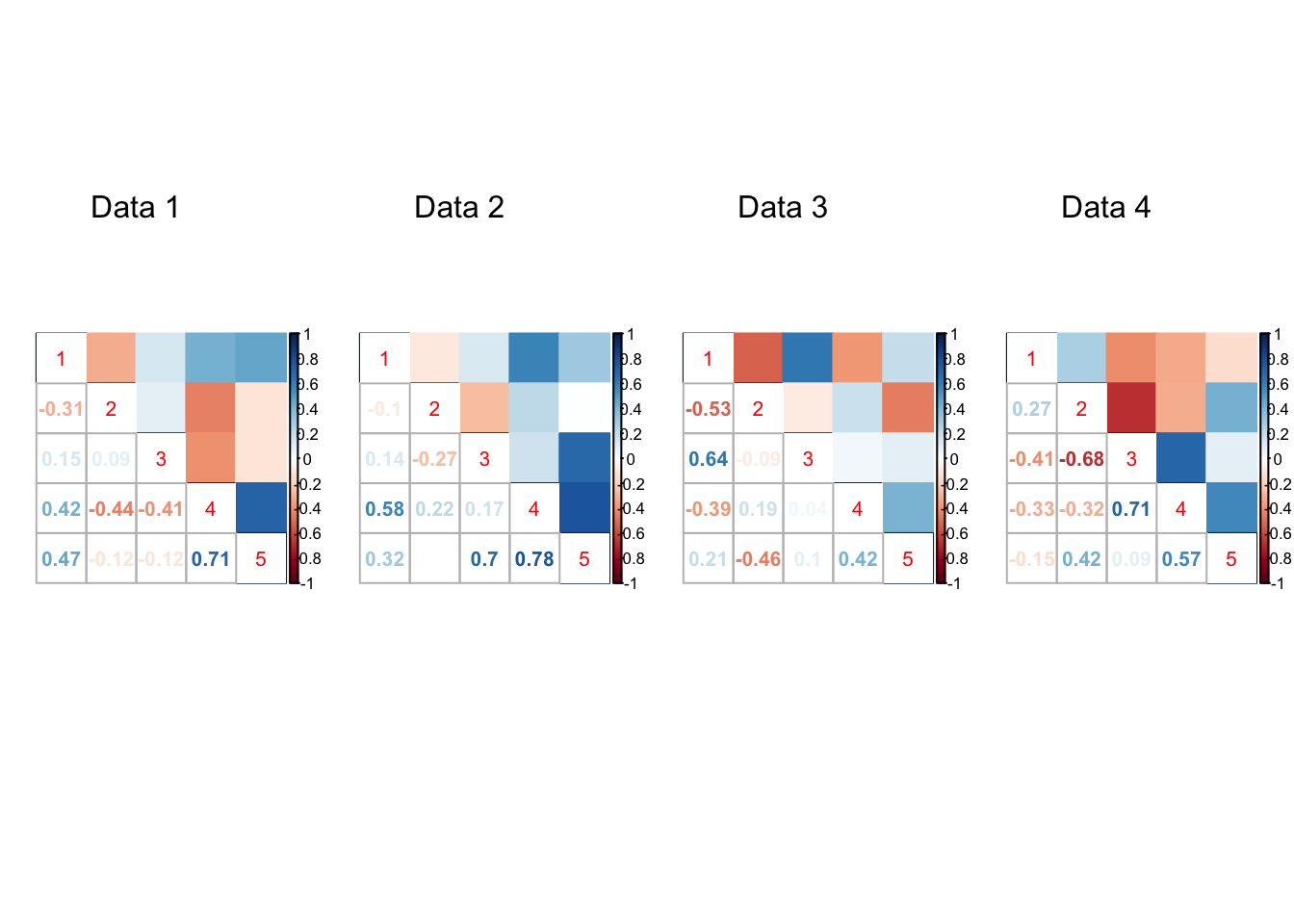
}par(mfrow=c(1,4))
for(t in 1:20){
corrplot::corrplot.mixed(result[[t]]$V.true, upper='color',cl.lim=c(-1,1))
mtext(paste0('Data ', t), at=2.5, line=-5)
}
Expand here to see past versions of unnamed-chunk-7-1.png:
| Version | Author | Date |
|---|---|---|
| 67990e8 | zouyuxin | 2018-08-21 |
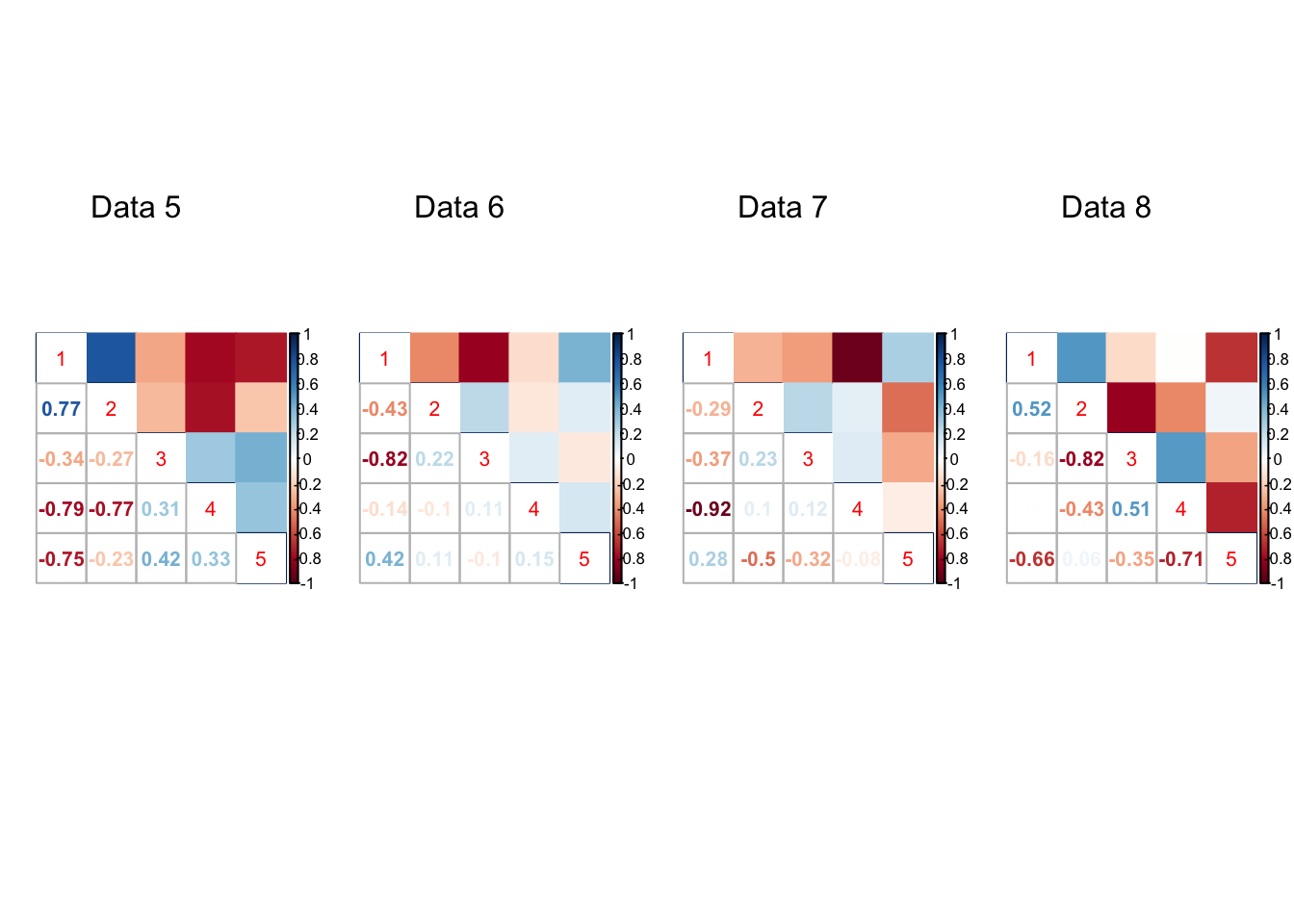
Expand here to see past versions of unnamed-chunk-7-2.png:
| Version | Author | Date |
|---|---|---|
| 67990e8 | zouyuxin | 2018-08-21 |
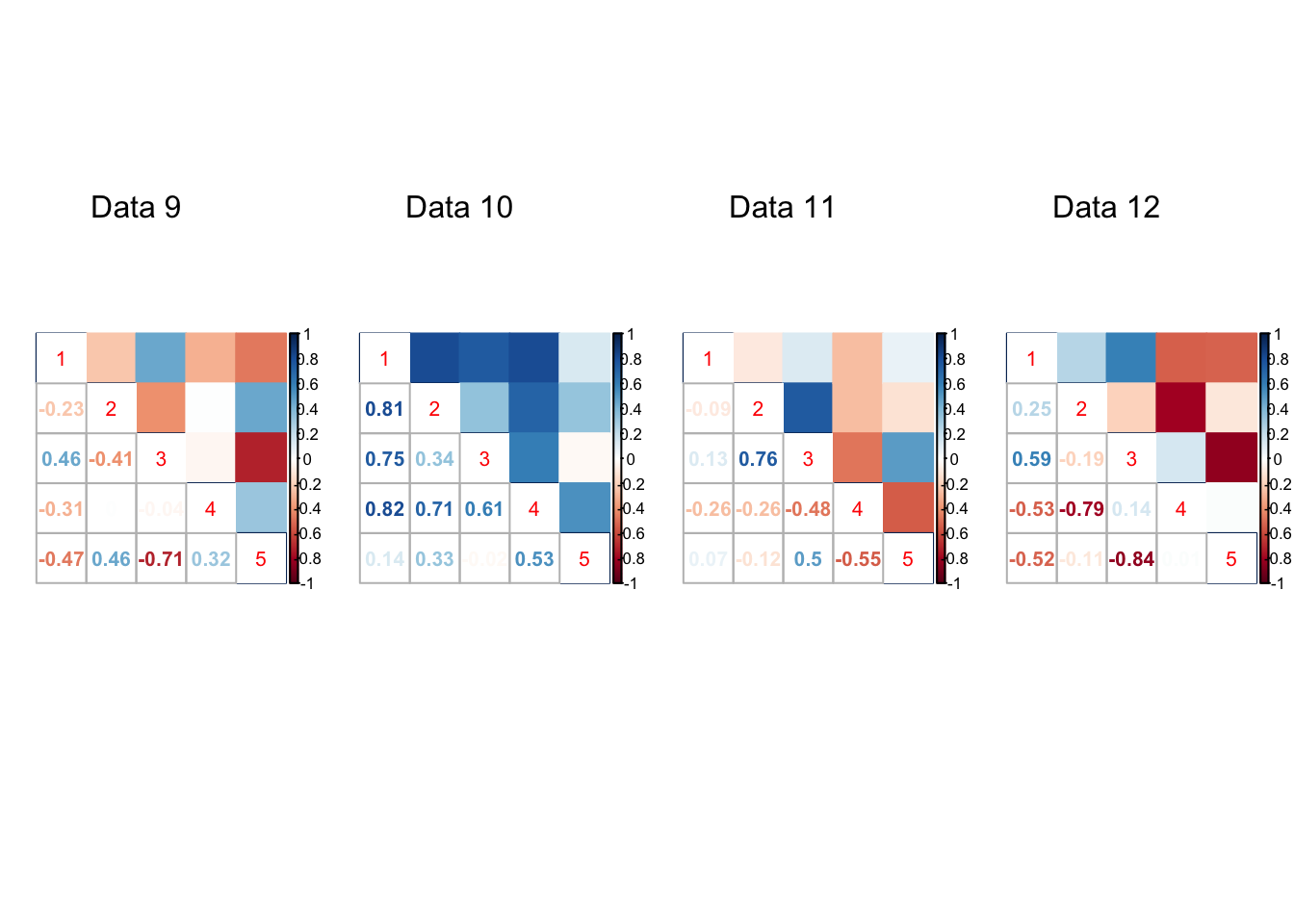
Expand here to see past versions of unnamed-chunk-7-3.png:
| Version | Author | Date |
|---|---|---|
| 67990e8 | zouyuxin | 2018-08-21 |
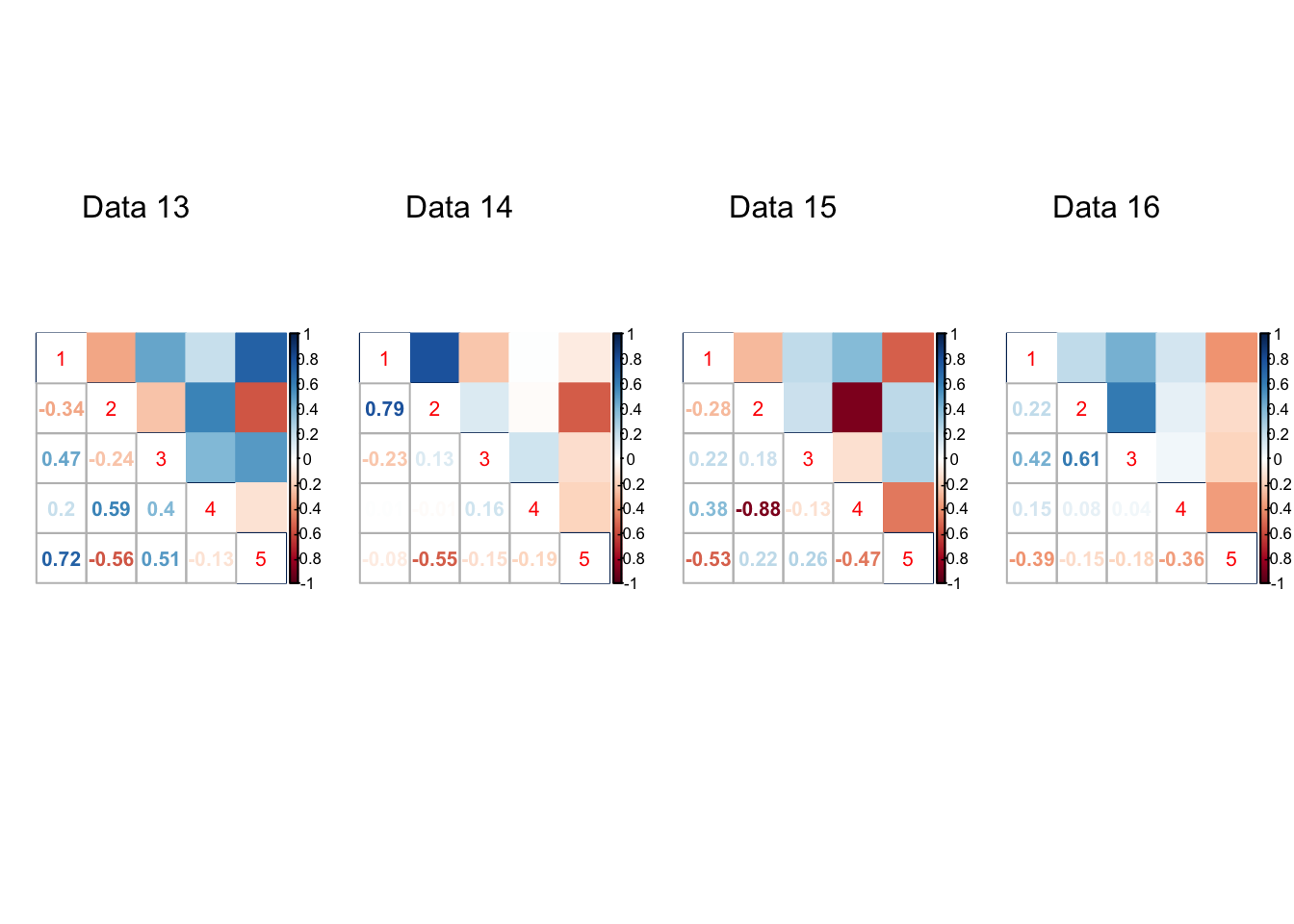
Expand here to see past versions of unnamed-chunk-7-4.png:
| Version | Author | Date |
|---|---|---|
| 67990e8 | zouyuxin | 2018-08-21 |
Expand here to see past versions of unnamed-chunk-7-5.png:
| Version | Author | Date |
|---|---|---|
| 67990e8 | zouyuxin | 2018-08-21 |
EM_res = c(); Trunc_res = c()
for(t in 1:20){
err = norm(result[[t]]$V.em$V - result[[t]]$V.true, type='F')
loglik = get_loglik(result[[t]]$model.em)
sig = length(get_significant_results(result[[t]]$model.em))
EM_res = rbind(EM_res, c(err, loglik, sig))
err = norm(result[[t]]$V.trunc - result[[t]]$V.true, type='F')
loglik = get_loglik(result[[t]]$model.trunc)
sig = length(get_significant_results(result[[t]]$model.trunc))
Trunc_res = rbind(Trunc_res, c(err, loglik, sig))
}mash results based on EM estimated V
colnames(EM_res) = c('F.error', 'loglik', '# significance')
EM_res %>% kable() %>% kable_styling()| F.error | loglik | # significance |
|---|---|---|
| 0.1420401 | -6233.044 | 0 |
| 0.1139972 | -4937.284 | 0 |
| 0.1508036 | -6045.184 | 0 |
| 0.0598159 | -5634.970 | 0 |
| 0.0803792 | -4498.151 | 2 |
| 0.1383140 | -5844.997 | 0 |
| 0.1223280 | -5527.147 | 3 |
| 0.0926035 | -4407.713 | 0 |
| 0.1893671 | -6227.554 | 0 |
| 0.1369380 | -4719.186 | 0 |
| 0.0697218 | -4974.055 | 0 |
| 0.0922341 | -5075.397 | 0 |
| 0.0588698 | -5705.655 | 0 |
| 0.1365943 | -5494.810 | 0 |
| 0.1478870 | -5734.789 | 0 |
| 0.1017556 | -6631.884 | 0 |
| 0.0838991 | -5504.995 | 0 |
| 0.0368434 | -5362.641 | 0 |
| 0.1262740 | -5736.854 | 0 |
| 0.1346086 | -4258.128 | 0 |
mash results based on original estimated V
colnames(Trunc_res) = c('F.error', 'loglik', '# significance')
Trunc_res %>% kable() %>% kable_styling()| F.error | loglik | # significance |
|---|---|---|
| 0.2812578 | -6263.074 | 0 |
| 0.2554575 | -4976.046 | 0 |
| 0.2110287 | -6082.465 | 0 |
| 0.2557471 | -5680.419 | 0 |
| 0.3482540 | -4581.797 | 0 |
| 0.2093880 | -5876.652 | 0 |
| 0.3888612 | -5557.576 | 0 |
| 0.4217467 | -4493.518 | 0 |
| 0.3356877 | -6272.532 | 0 |
| 0.2516301 | -4786.389 | 0 |
| 0.2615190 | -5022.811 | 0 |
| 0.2619482 | -5134.581 | 0 |
| 0.3126799 | -5762.630 | 0 |
| 0.1615910 | -5523.273 | 0 |
| 0.2107430 | -5768.520 | 0 |
| 0.3014296 | -6652.888 | 0 |
| 0.2136931 | -5566.659 | 0 |
| 0.2679966 | -5403.671 | 0 |
| 0.2831679 | -5794.189 | 0 |
| 0.2645471 | -4312.278 | 0 |
Investigate Data 7
data = result[[7]]$data
V.true = result[[7]]$V.true
V.em = result[[7]]$V.em
model.em = result[[7]]$model.em
barplot(get_estimated_pi(model.em), las=2, cex.names = 0.7)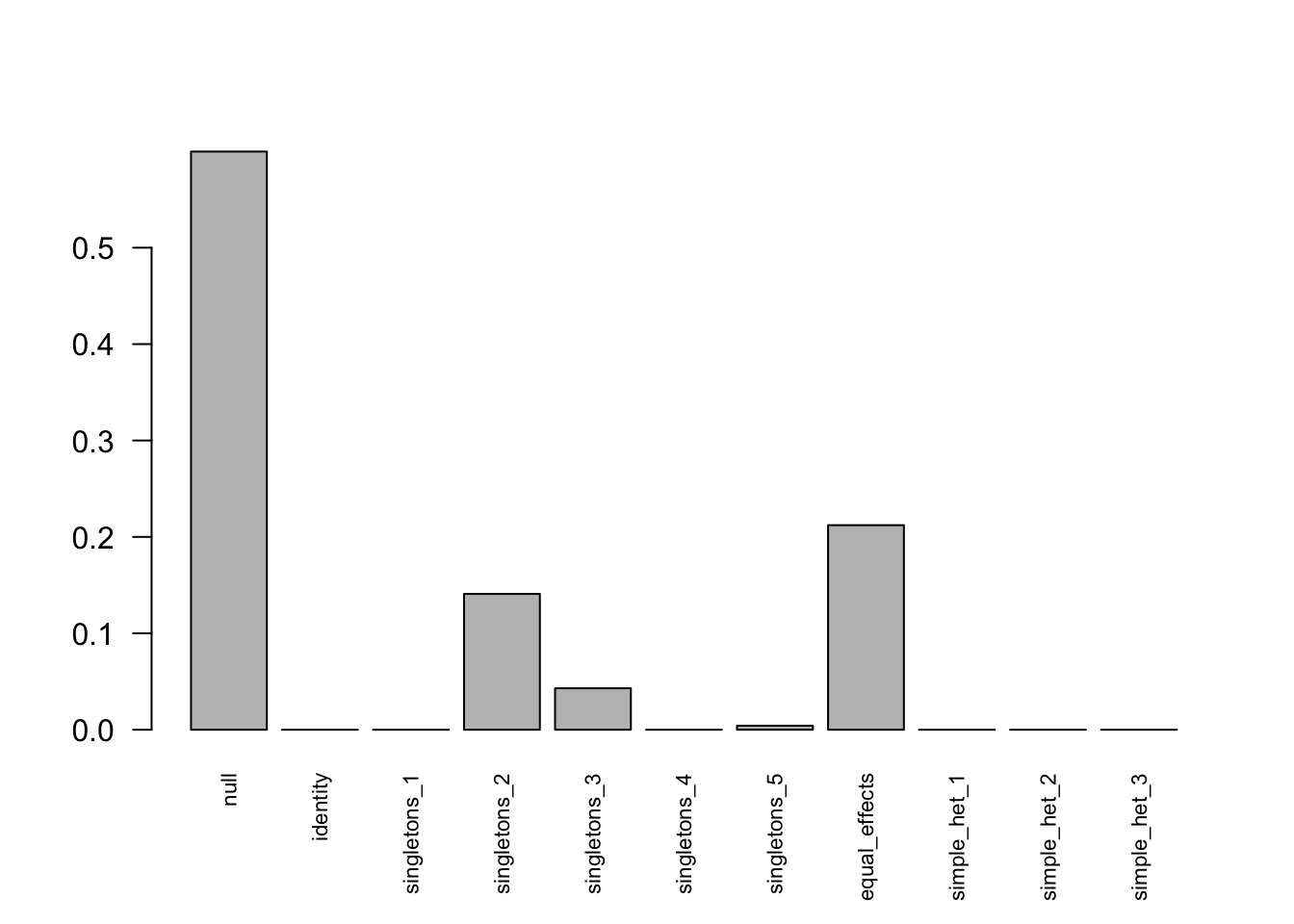
Expand here to see past versions of unnamed-chunk-11-1.png:
| Version | Author | Date |
|---|---|---|
| 67990e8 | zouyuxin | 2018-08-21 |
samp = result[[7]]$sample
D1 = lapply(data, function(m) m[samp,])
D2 = lapply(data, function(m) m[-samp,])Estimate V on D1
m.data1 = mash_set_data(Bhat = D1$Bhat, Shat = D1$Shat)
U.c = cov_canonical(m.data1)
Vhat.em = estimateV(m.data1, U.c, init_rho = c(-0.5,0,0.5), tol=1e-4, optmethod='em2')Estimate mixture proportions on D2
m.data2 = mash_set_data(Bhat = D2$Bhat, Shat = D2$Shat, V = Vhat.em$V)
m.model.split = mash(m.data2, U.c, outputlevel = 1) - Computing 1000 x 141 likelihood matrix.
- Likelihood calculations took 0.01 seconds.
- Fitting model with 141 mixture components.
- Model fitting took 0.10 seconds.Estimate posteior on D1
m.data1.em = mash_set_data(Bhat = D1$Bhat, Shat = D1$Shat, V = Vhat.em$V)
m.model.split$result = mash_compute_posterior_matrices(m.model.split, m.data1.em)The # significant samples
length(get_significant_results(m.model.split))[1] 2Session information
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] kableExtra_0.9.0 knitr_1.20 mashr_0.2-11 ashr_2.2-10
loaded via a namespace (and not attached):
[1] Rcpp_0.12.18 highr_0.7 pillar_1.3.0
[4] compiler_3.5.1 git2r_0.23.0 plyr_1.8.4
[7] workflowr_1.1.1 R.methodsS3_1.7.1 R.utils_2.6.0
[10] iterators_1.0.10 tools_3.5.1 corrplot_0.84
[13] digest_0.6.15 viridisLite_0.3.0 tibble_1.4.2
[16] evaluate_0.11 lattice_0.20-35 pkgconfig_2.0.2
[19] rlang_0.2.2 Matrix_1.2-14 foreach_1.4.4
[22] rstudioapi_0.7 yaml_2.2.0 parallel_3.5.1
[25] mvtnorm_1.0-8 xml2_1.2.0 httr_1.3.1
[28] stringr_1.3.1 REBayes_1.3 hms_0.4.2
[31] rprojroot_1.3-2 grid_3.5.1 R6_2.2.2
[34] rmarkdown_1.10 rmeta_3.0 readr_1.1.1
[37] magrittr_1.5 whisker_0.3-2 scales_1.0.0
[40] backports_1.1.2 codetools_0.2-15 htmltools_0.3.6
[43] MASS_7.3-50 rvest_0.3.2 assertthat_0.2.0
[46] colorspace_1.3-2 stringi_1.2.4 Rmosek_8.0.69
[49] munsell_0.5.0 doParallel_1.0.11 pscl_1.5.2
[52] truncnorm_1.0-8 SQUAREM_2017.10-1 crayon_1.3.4
[55] R.oo_1.22.0 This reproducible R Markdown analysis was created with workflowr 1.1.1