UKBioBank FLASH
Yuxin Zou
2018-5-30
Last updated: 2018-06-04
Code version: 479d555
Loading required package: ashrUKBioBank Strong data
data = readRDS('../data/UKBioBank/StrongData.rds')Estimate se based on p values
# Adjust p value == 0
data$p[data$p == 0] = 1e-323
mash.data = mash_set_data(Bhat = data$beta, pval = data$p)Flash on centered Z
Z = mash.data$Bhat/mash.data$Shat
# column center
Z.center = apply(Z, 2, function(x) x - mean(x, na.rm = TRUE))flash.data = flash_set_data(Z.center)
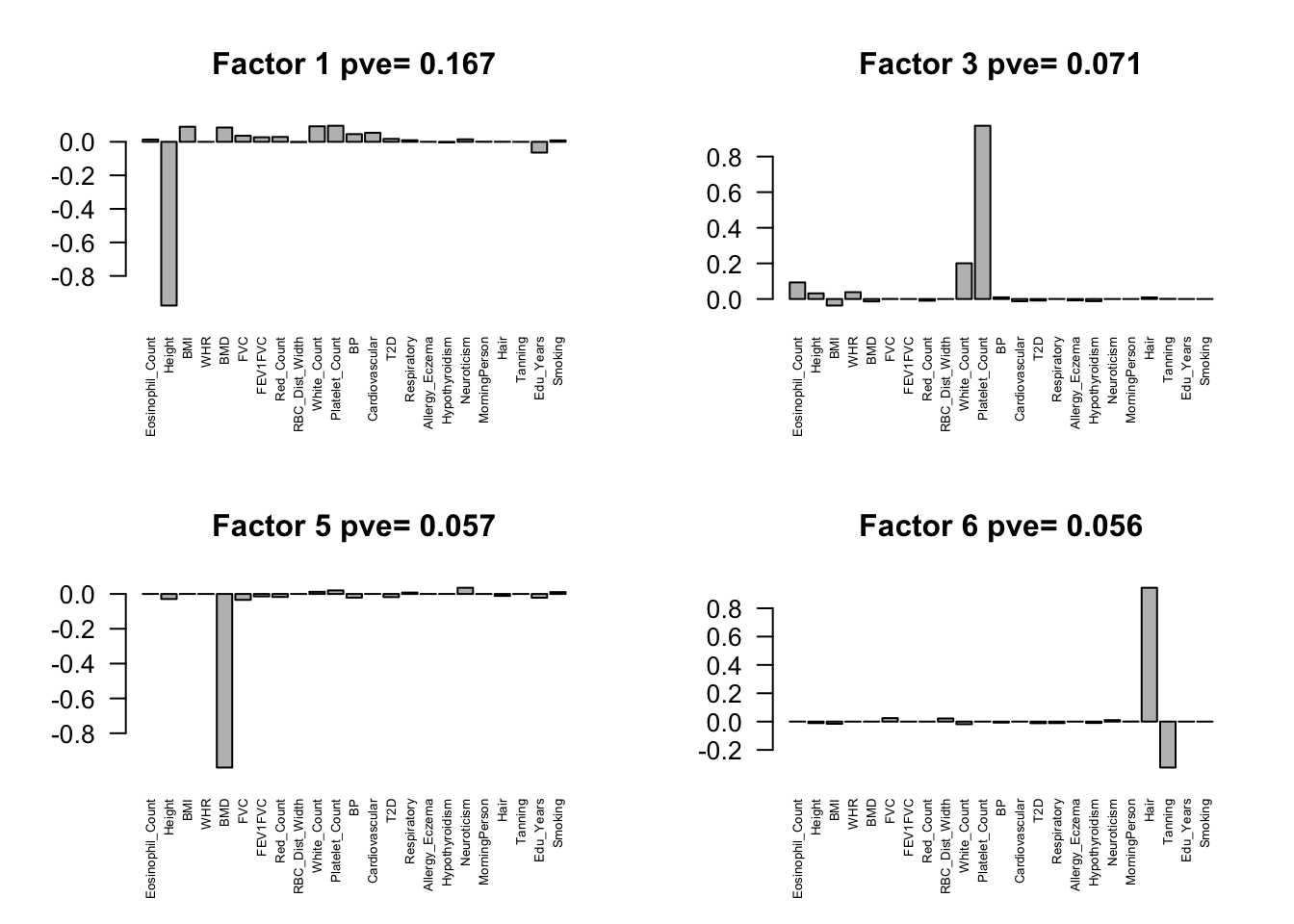
fmodel = flash(flash.data, greedy = TRUE, backfit = TRUE)fitting factor/loading 1fitting factor/loading 2fitting factor/loading 3fitting factor/loading 4fitting factor/loading 5fitting factor/loading 6fitting factor/loading 7fitting factor/loading 8fitting factor/loading 9fitting factor/loading 10fitting factor/loading 11fitting factor/loading 12fitting factor/loading 13fitting factor/loading 14fitting factor/loading 15fitting factor/loading 16fitting factor/loading 17fitting factor/loading 18fitting factor/loading 19Factors = flash_get_ldf(fmodel)$f
row.names(Factors) = colnames(Z)
pve.order = order(flash_get_pve(fmodel), decreasing = TRUE)
par(mfrow=c(2,2))
for(i in pve.order){
barplot(Factors[,i], main=paste0('Factor ',i, ' pve= ', round(flash_get_pve(fmodel)[i],3)), las=2, cex.names = 0.5)
}
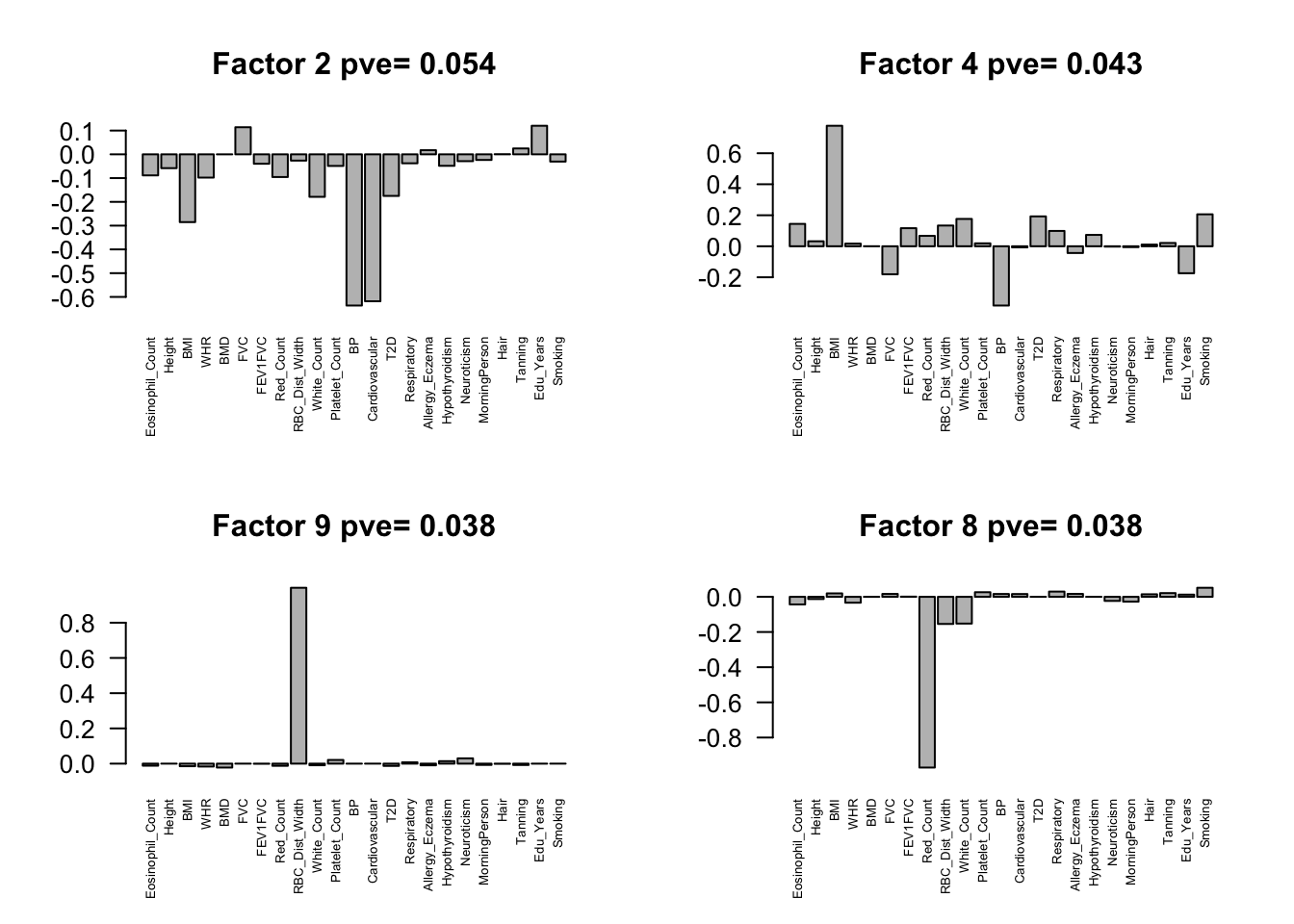
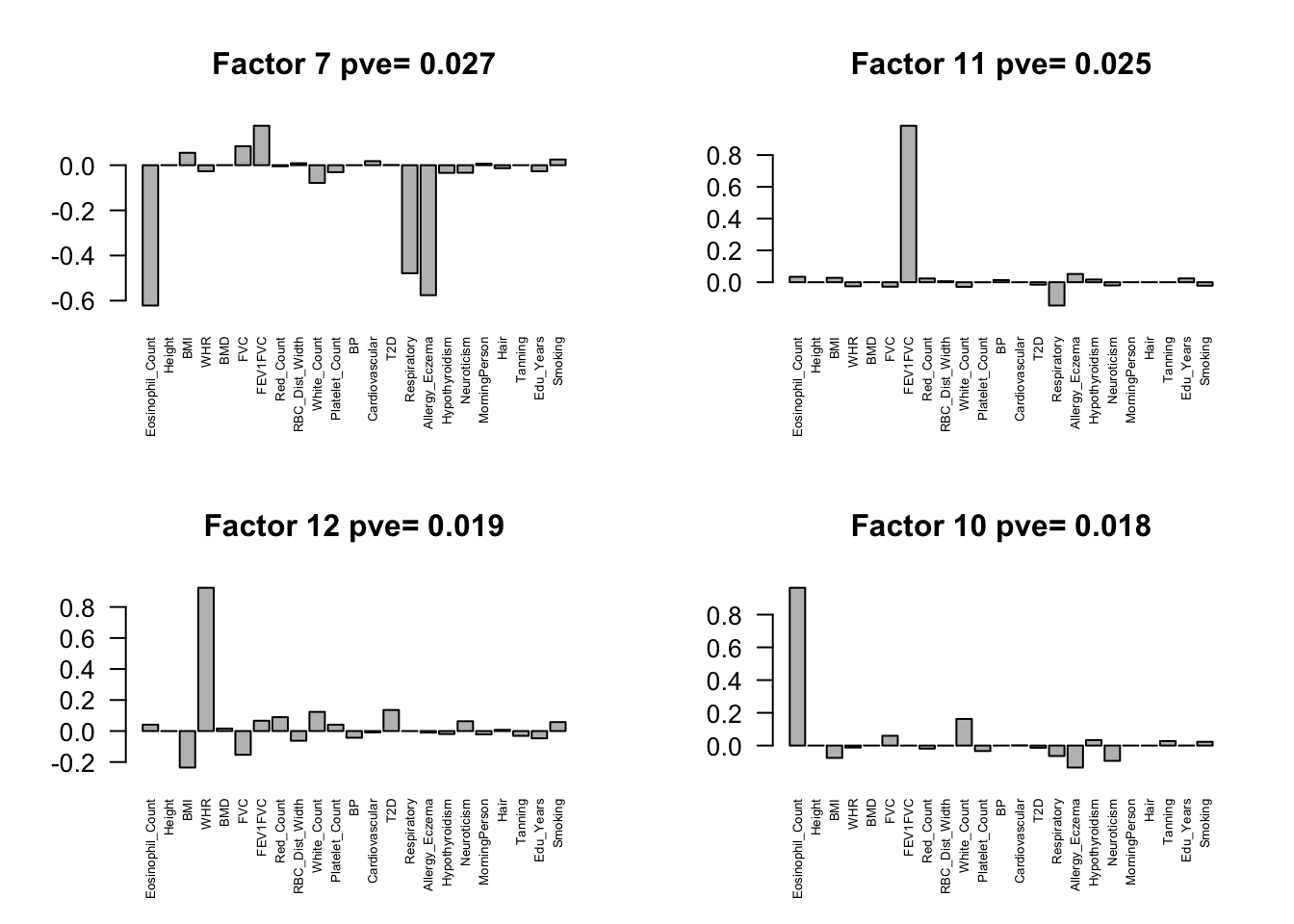
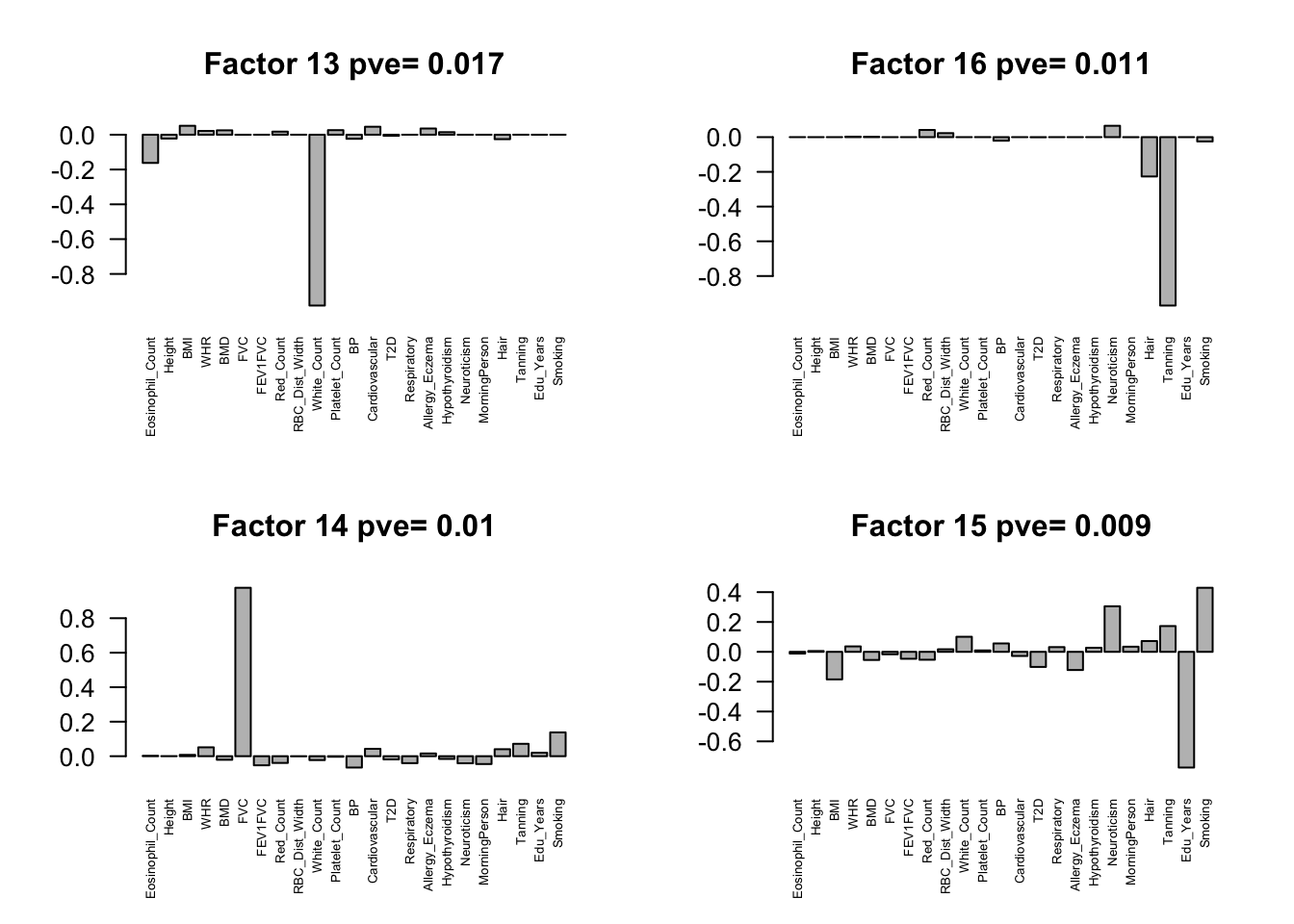
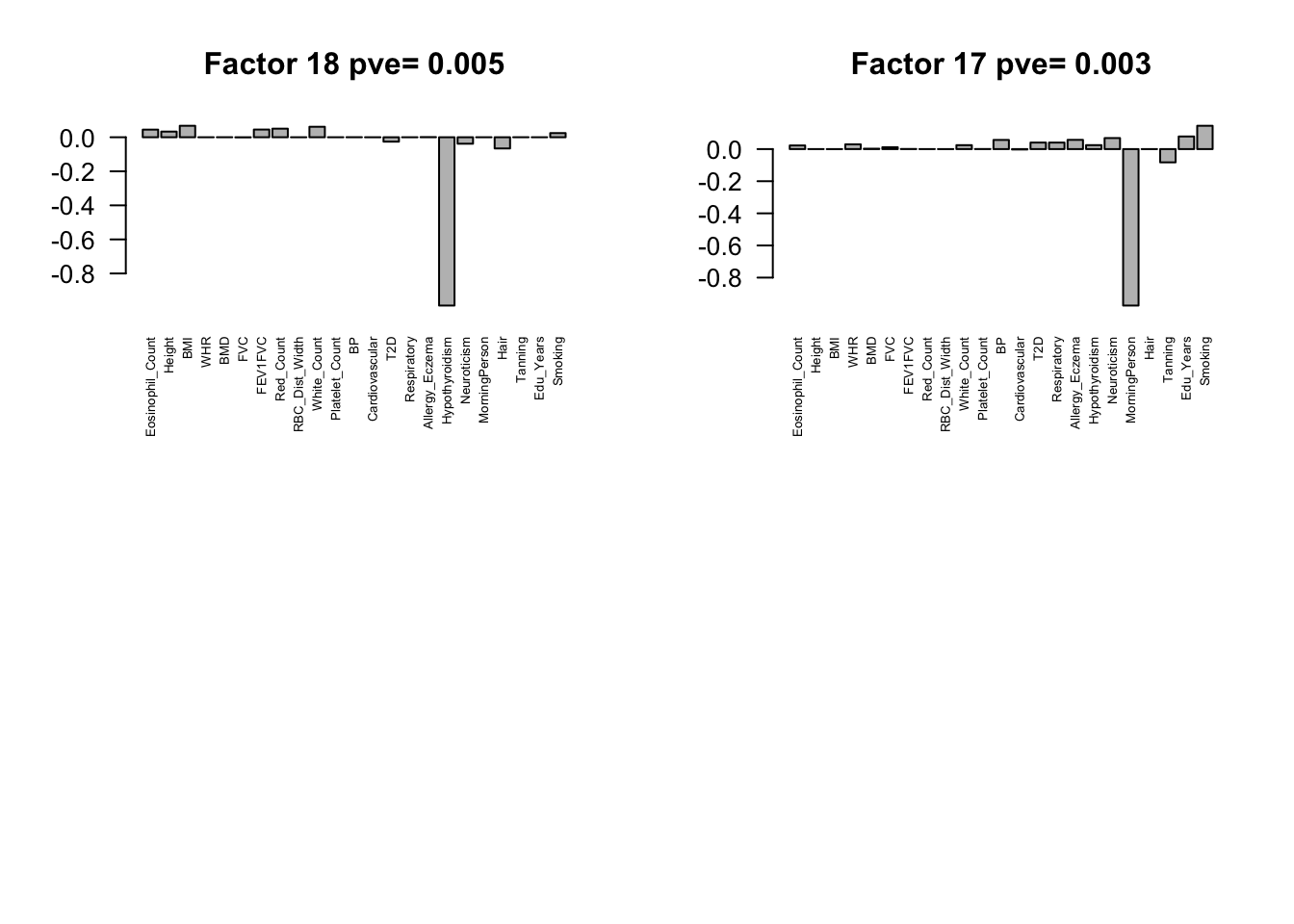
Flash again on the loading matrix
loading = fmodel$EL[,1:18]
colnames(loading) = paste0('Factor',seq(1,18))
flash.loading = flash_set_data(loading)
flmodel = flash(flash.loading, greedy = TRUE, backfit = TRUE)fitting factor/loading 1fitting factor/loading 2fitting factor/loading 3fitting factor/loading 4fitting factor/loading 5fitting factor/loading 6fitting factor/loading 7fitting factor/loading 8fitting factor/loading 9fitting factor/loading 10fitting factor/loading 11fitting factor/loading 12fitting factor/loading 13fitting factor/loading 14fitting factor/loading 15fitting factor/loading 16fitting factor/loading 17fitting factor/loading 18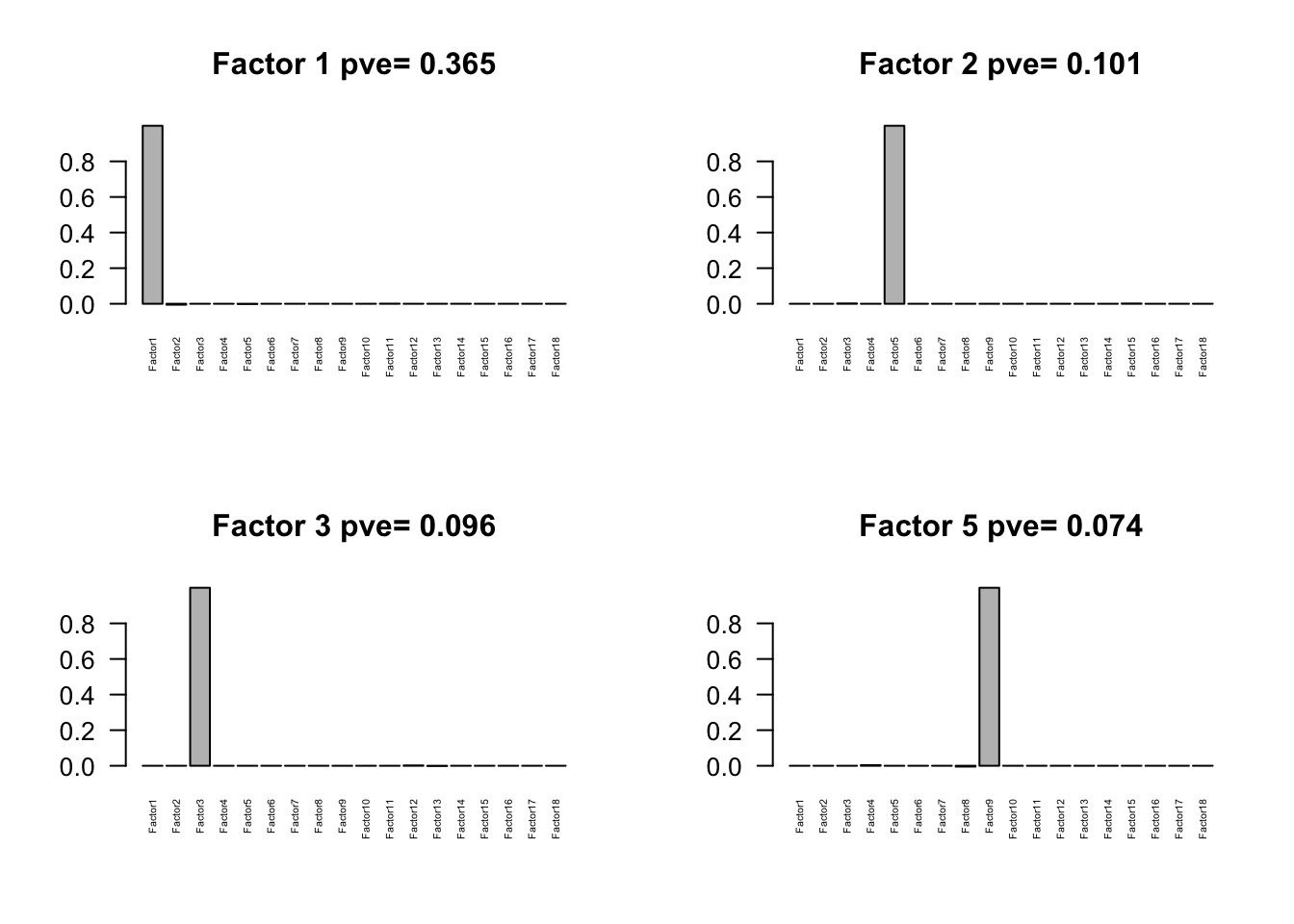
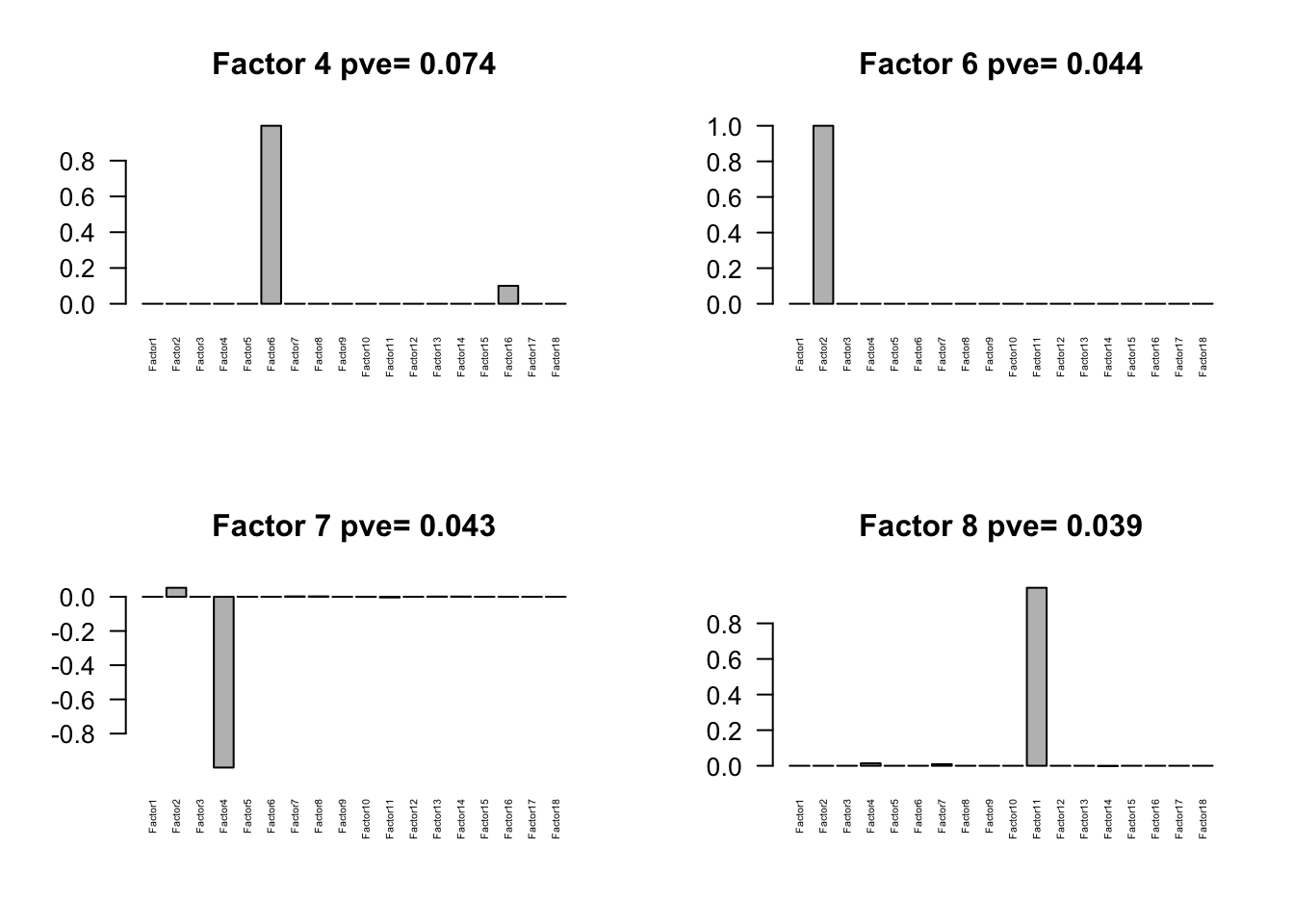
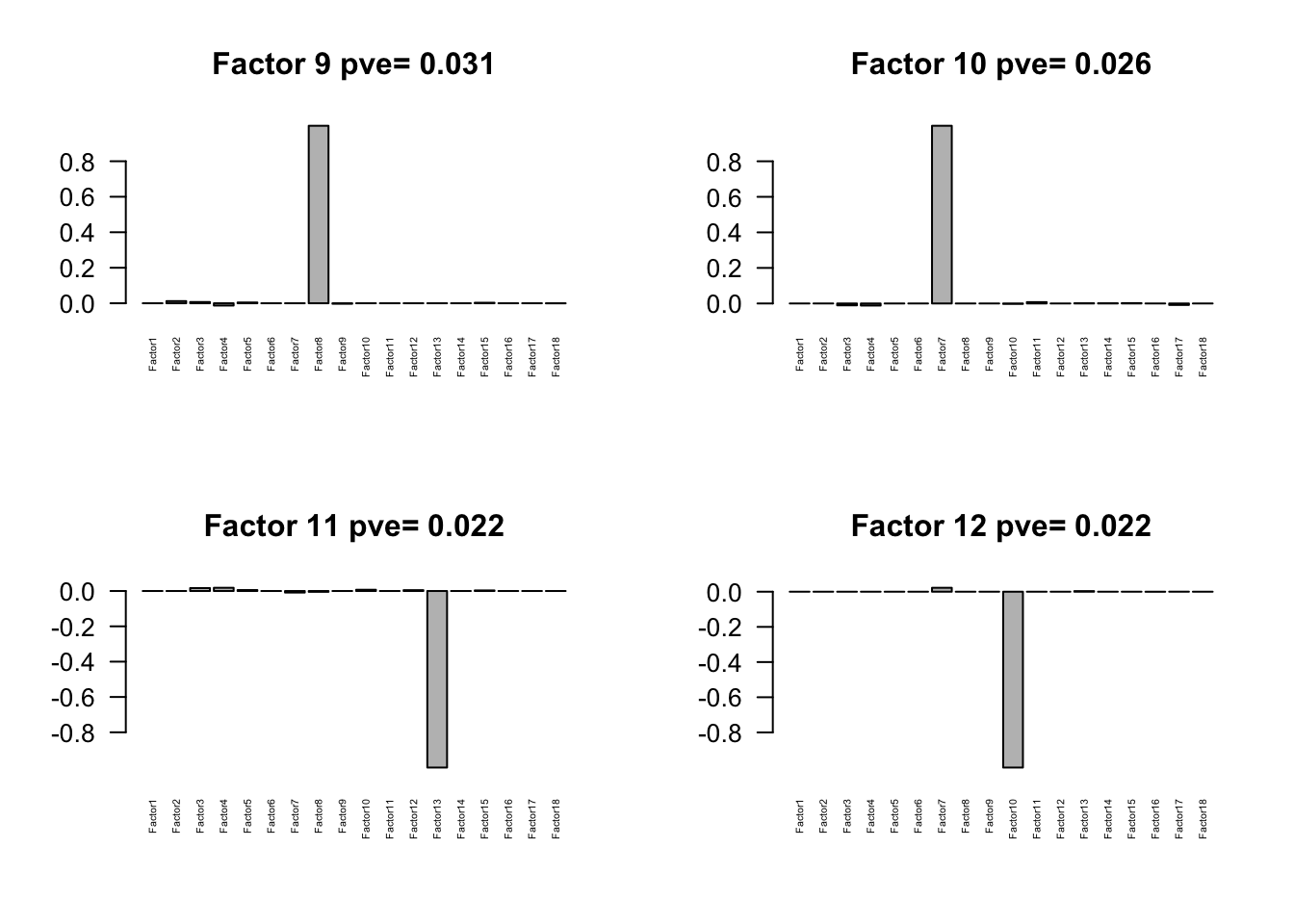

Session information
sessionInfo()R version 3.4.4 (2018-03-15)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS High Sierra 10.13.4
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.4/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] mashr_0.2-10 ashr_2.2-7 flashr_0.5-8
loaded via a namespace (and not attached):
[1] Rcpp_0.12.17 compiler_3.4.4 pillar_1.2.2
[4] git2r_0.21.0 plyr_1.8.4 iterators_1.0.9
[7] tools_3.4.4 digest_0.6.15 evaluate_0.10.1
[10] tibble_1.4.2 gtable_0.2.0 lattice_0.20-35
[13] rlang_0.2.0 Matrix_1.2-14 foreach_1.4.4
[16] yaml_2.1.19 parallel_3.4.4 mvtnorm_1.0-7
[19] ebnm_0.1-11 stringr_1.3.0 knitr_1.20
[22] rprojroot_1.3-2 grid_3.4.4 rmarkdown_1.9
[25] rmeta_3.0 ggplot2_2.2.1 magrittr_1.5
[28] backports_1.1.2 scales_0.5.0 codetools_0.2-15
[31] htmltools_0.3.6 MASS_7.3-50 assertthat_0.2.0
[34] softImpute_1.4 colorspace_1.3-2 stringi_1.2.2
[37] lazyeval_0.2.1 pscl_1.5.2 doParallel_1.0.11
[40] munsell_0.4.3 truncnorm_1.0-8 SQUAREM_2017.10-1This R Markdown site was created with workflowr