n \(>\) p
We run similations with n > p. Let n = 1000, p=500. The susie model captures the true effects in all cases below.
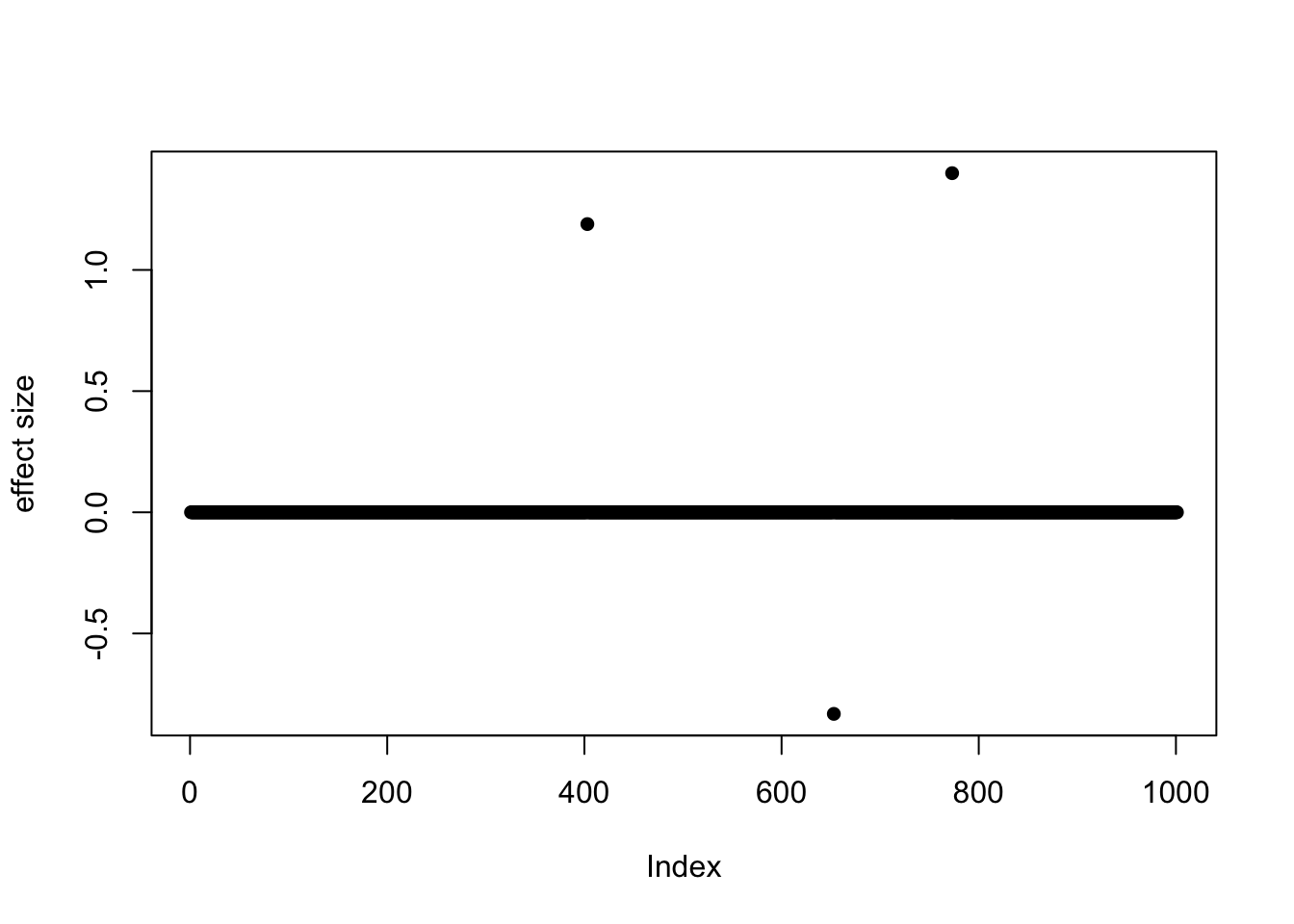
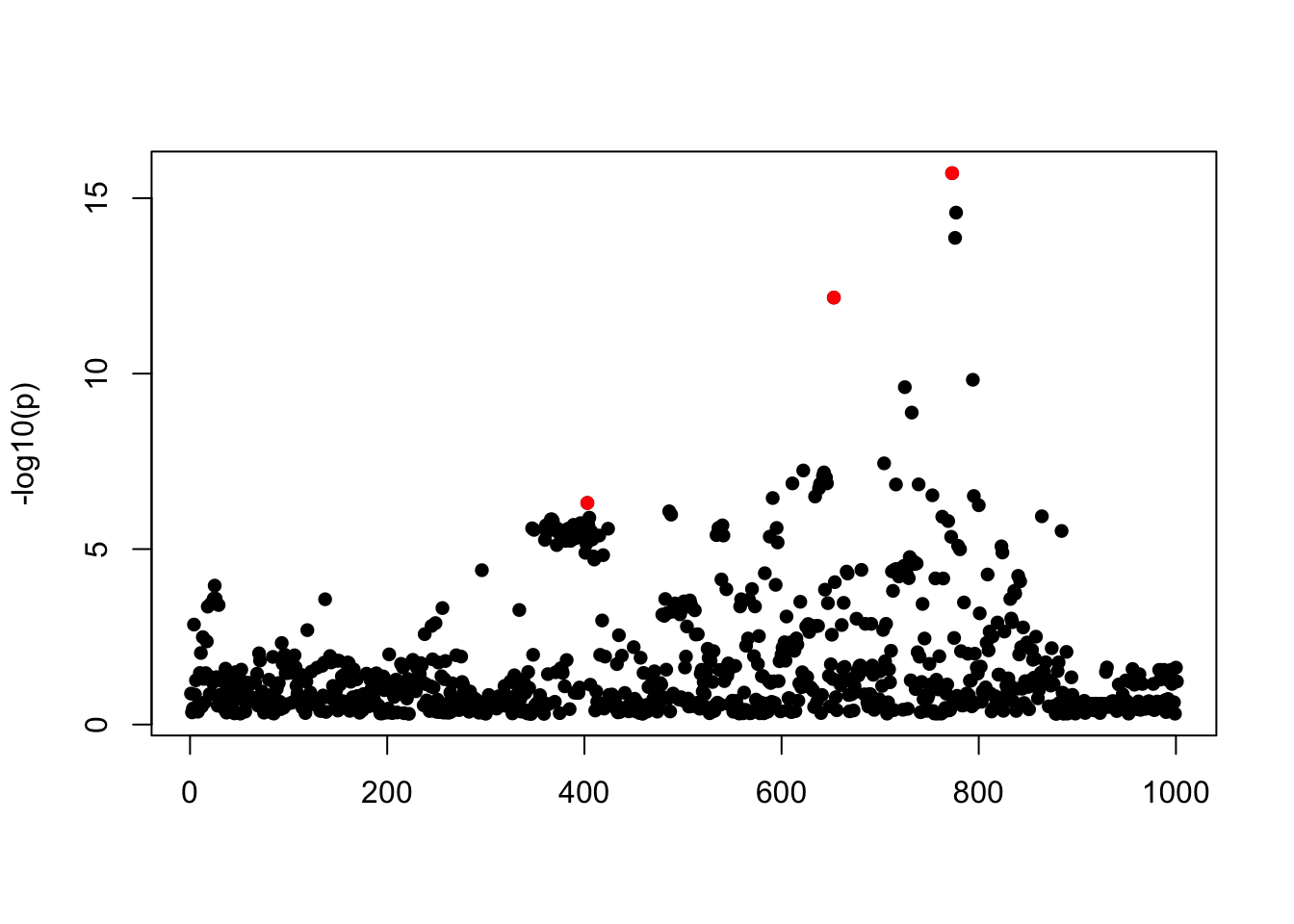
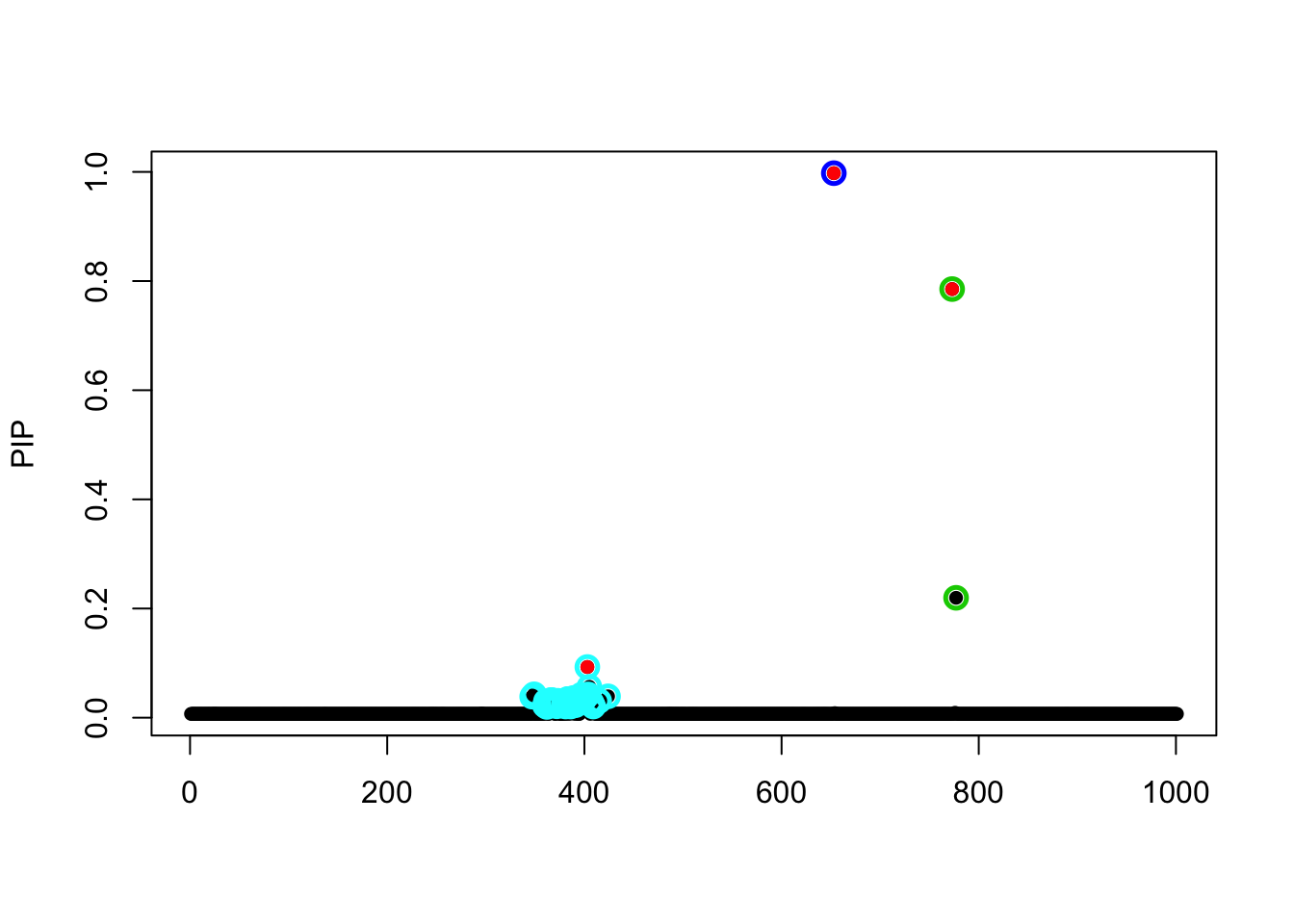
- Case 1: L=1. The true effect is b200. The response y is simulated from the specified bernoulli model without intercept, which means the number of case-control is well-balanced. The susie model captures the causal effect.
set.seed(1)
n = 1000
p = 500
X = matrix(rnorm(n*p, 0, 1), nrow = n, ncol = p)
R = cor(X)
beta_true = rep(0, p)
beta_true[200] = 1
Y = rbinom(n, 1, exp(X %*% beta_true) / (1 + exp(X %*% beta_true)))
z = numeric(p)
for(i in 1:p){
z[i] = summary(glm(Y~X[,i], family = 'binomial'))$coef[2,3]
}
susie_plot(z, y='z', b=beta_true)

Expand here to see past versions of unnamed-chunk-4-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
|
213b965
|
zouyuxin
|
2018-12-04
|
fit_z = susieR::susie_z(z, R, min_abs_corr = 0)
susie_plot(fit_z, y="PIP", b=beta_true)

Expand here to see past versions of unnamed-chunk-6-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
|
cb3f4e5
|
zouyuxin
|
2018-12-05
|
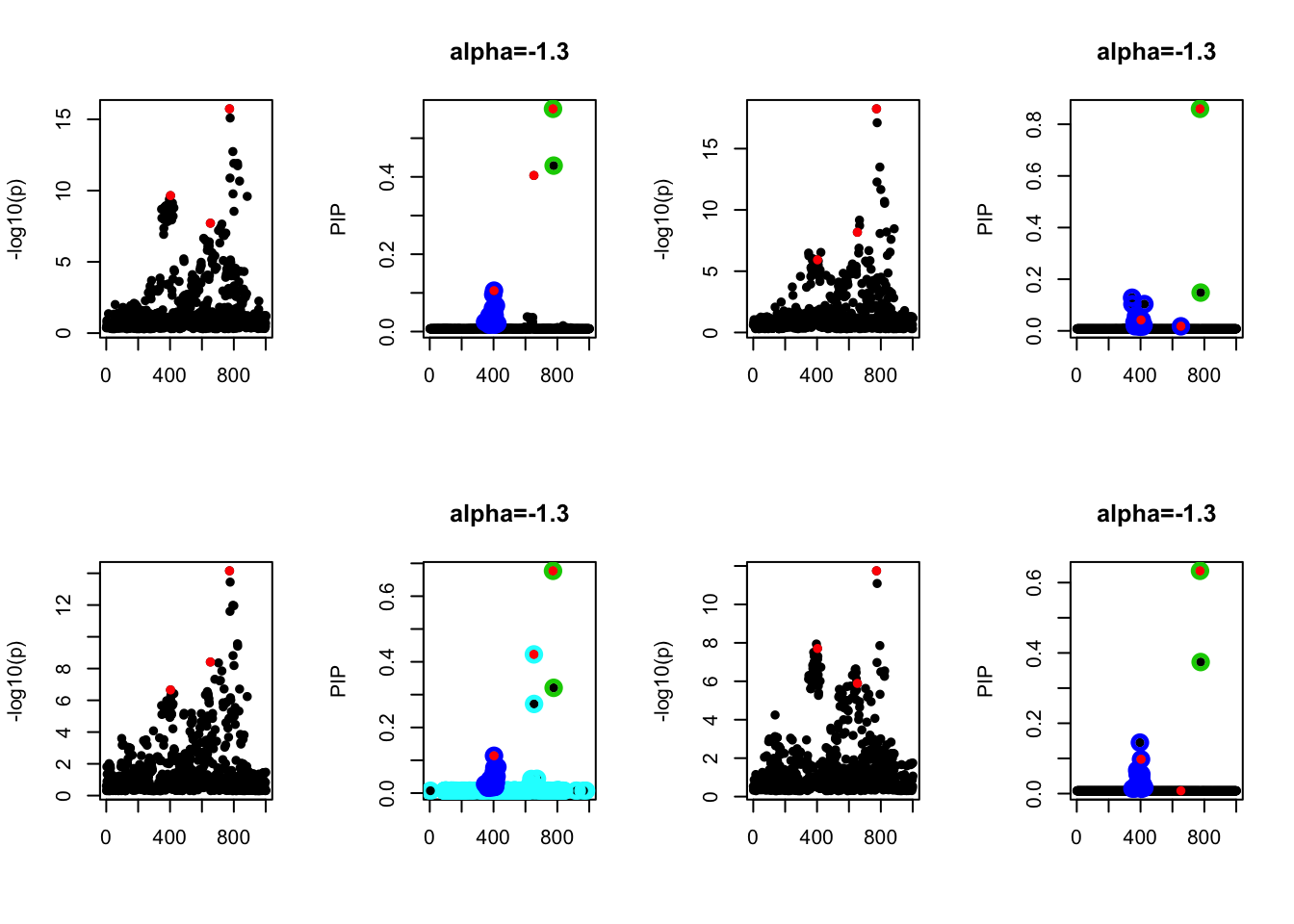
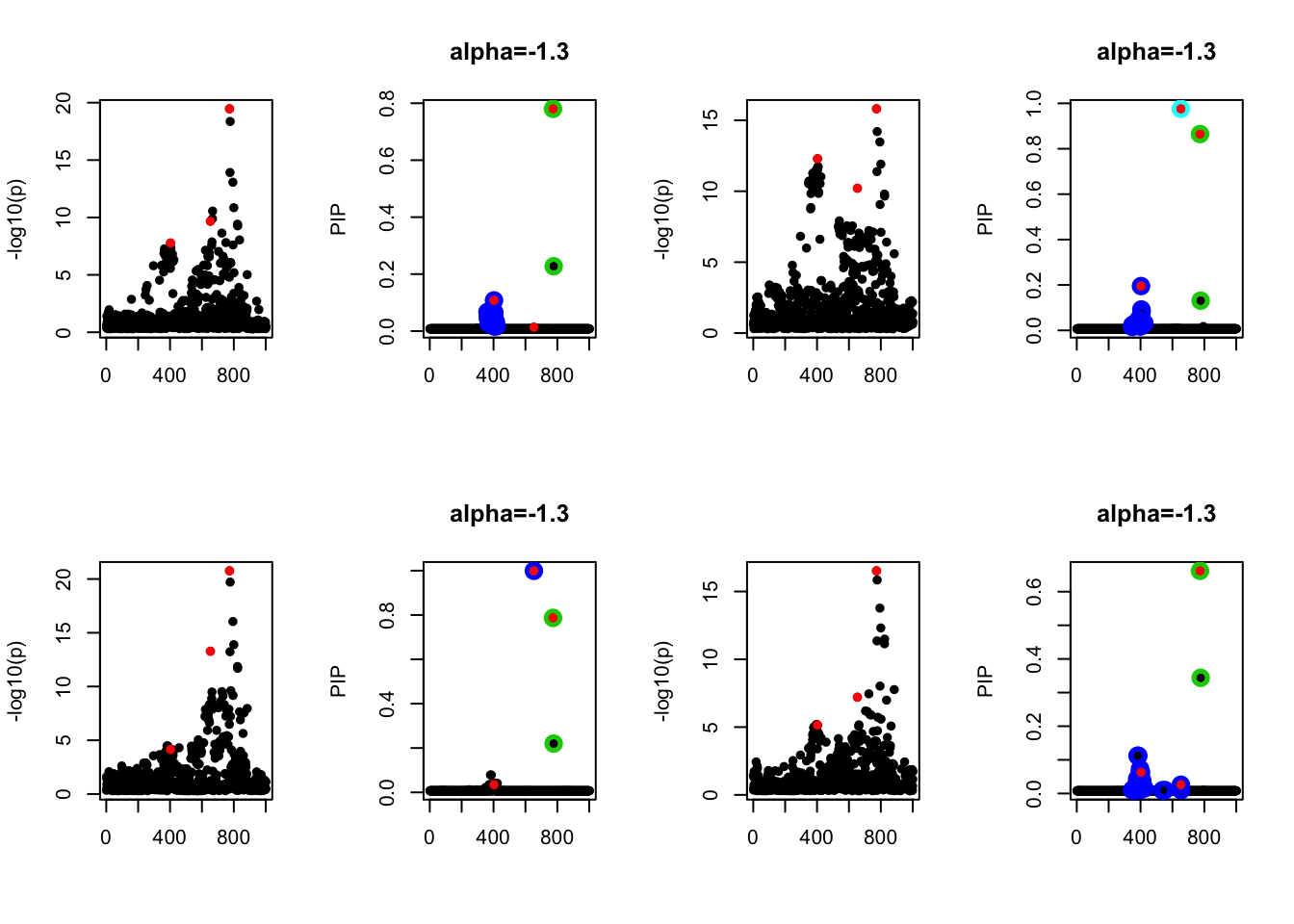
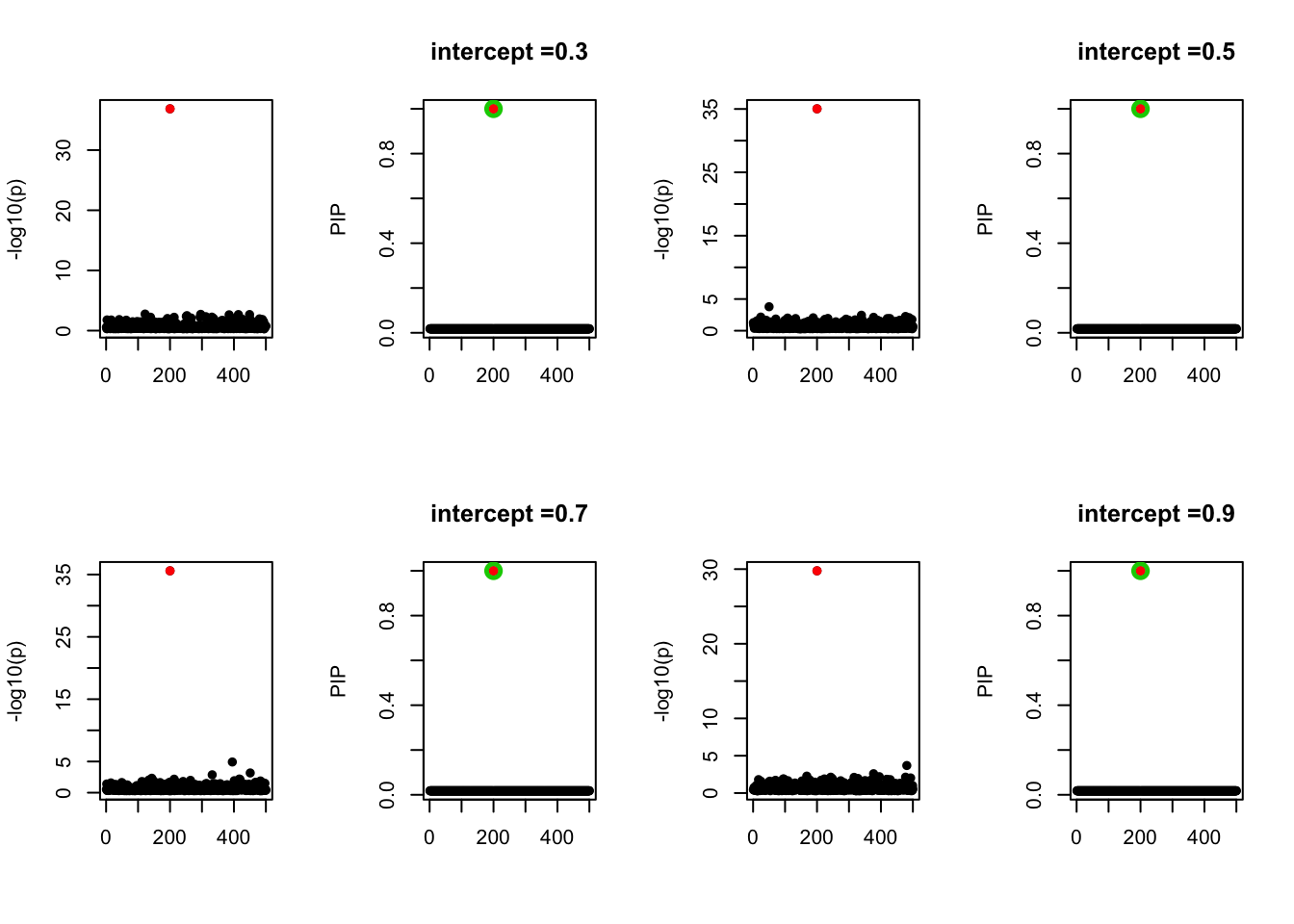
- Case 2: The response y is simulated from the specified bernoulli model with intercept from -1.3 to 1.3. When the intercept is not zero, the case-control is not balanced. The susie model captures the causal effect.
par(mfrow=c(2, 4))
alpha = seq(-1.3,1.3,by=0.2)
set.seed(1)
for(a in alpha){
Y = rbinom(n, 1, exp(a+X %*% beta_true) / (1 + exp(a+X %*% beta_true)))
z = numeric(p)
for(i in 1:p){
z[i] = summary(glm(Y~X[,i], family = 'binomial'))$coef[2,3]
}
susie_plot(z, y='z', b=beta_true)
fit_z = susieR::susie_z(z, R, min_abs_corr = 0)
susie_plot(fit_z, y="PIP", b=beta_true, main=paste0('intercept =', a))
}

Expand here to see past versions of unnamed-chunk-7-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
|
213b965
|
zouyuxin
|
2018-12-04
|
Expand here to see past versions of unnamed-chunk-7-2.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
Expand here to see past versions of unnamed-chunk-7-3.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
Expand here to see past versions of unnamed-chunk-7-4.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
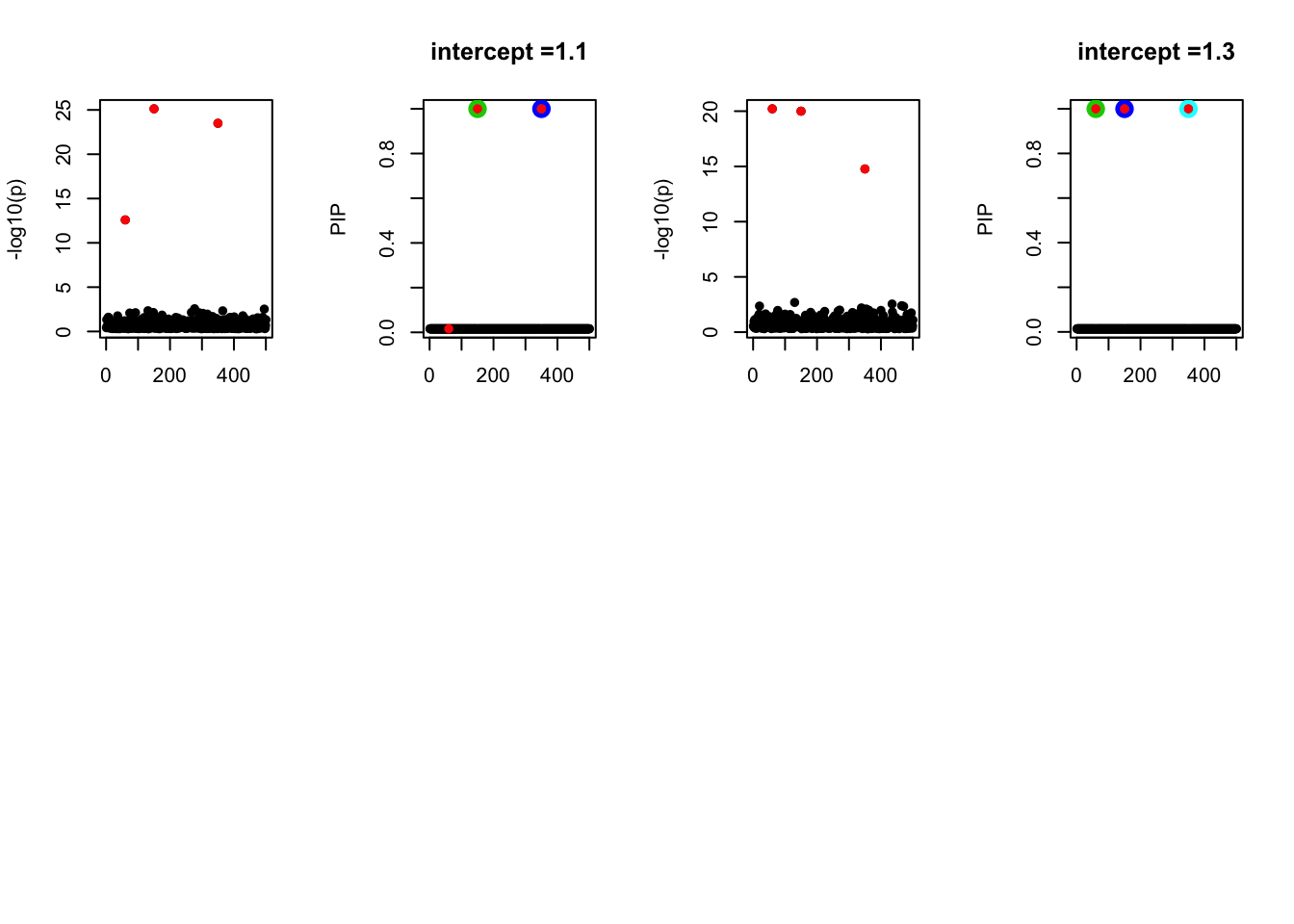
- Case 3: We increase the number of true effects to be 3. The true effects are b60, b150, b350. The response y is simulated from the specified bernoulli model without intercept. The susie model captures 2 causal effects.
beta_true = rep(0, p)
beta_true[c(60,350)] = 1
beta_true[150] = -1
set.seed(1)
Y = rbinom(n, 1, exp(X %*% beta_true) / (1 + exp(X %*% beta_true)))
z = numeric(p)
for(i in 1:p){
z[i] = summary(glm(Y~X[,i], family = 'binomial'))$coef[2,3]
}
susie_plot(z, y='z', b=beta_true)

Expand here to see past versions of unnamed-chunk-8-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
|
cb3f4e5
|
zouyuxin
|
2018-12-05
|
fit_z = susieR::susie_z(z, R, min_abs_corr = 0)
susie_plot(fit_z, y="PIP", b=beta_true)

Expand here to see past versions of unnamed-chunk-9-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
|
cb3f4e5
|
zouyuxin
|
2018-12-05
|
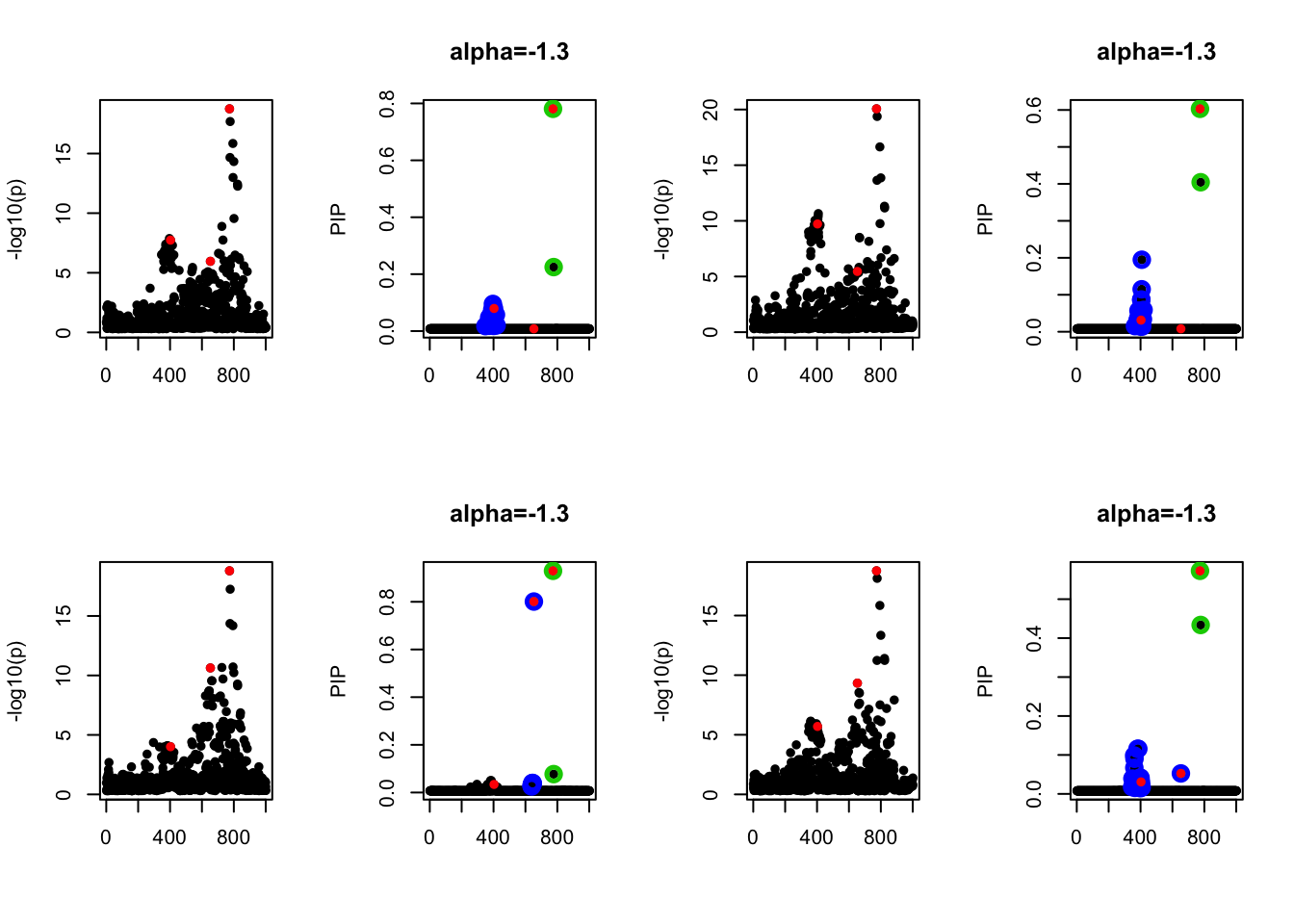
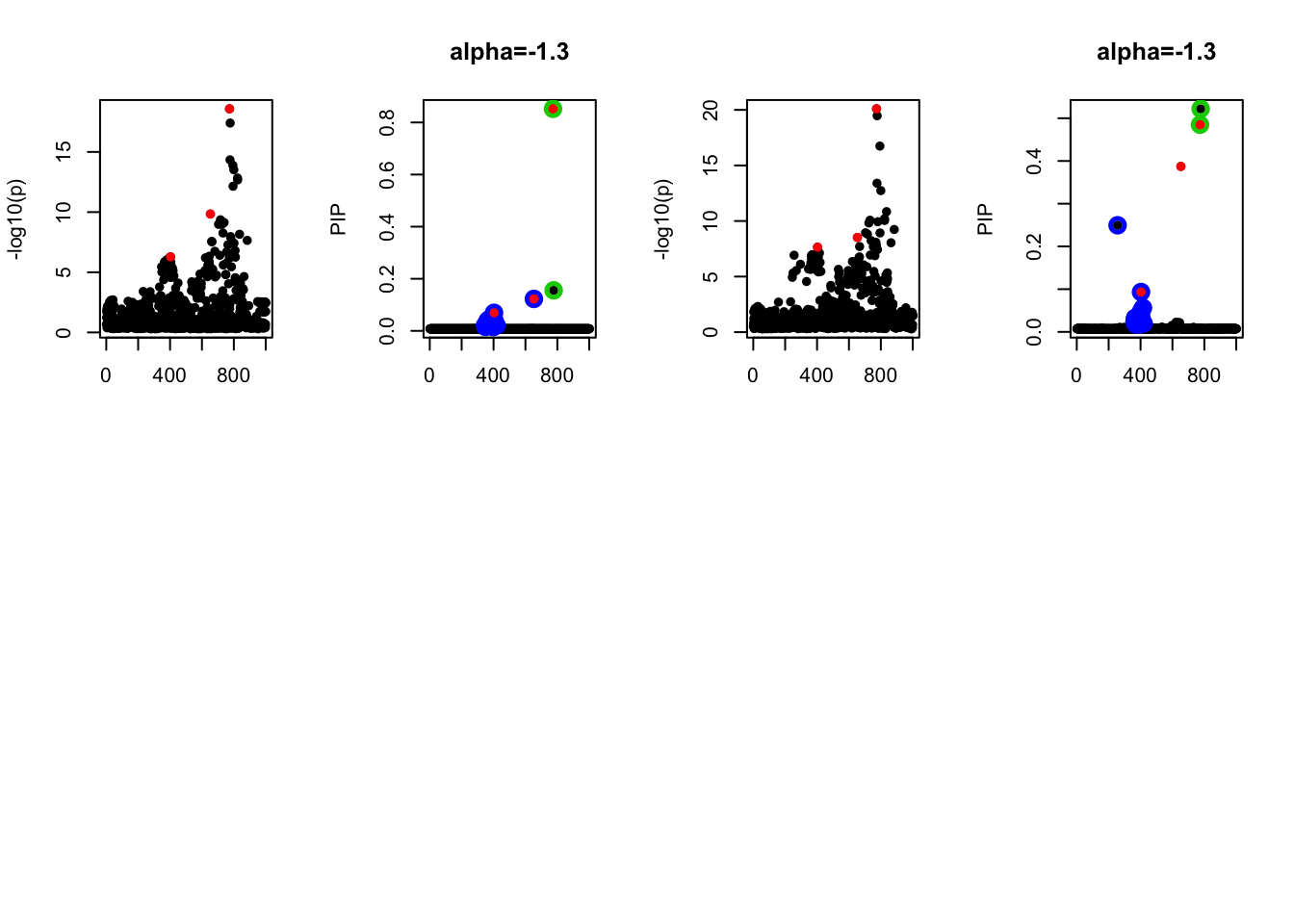
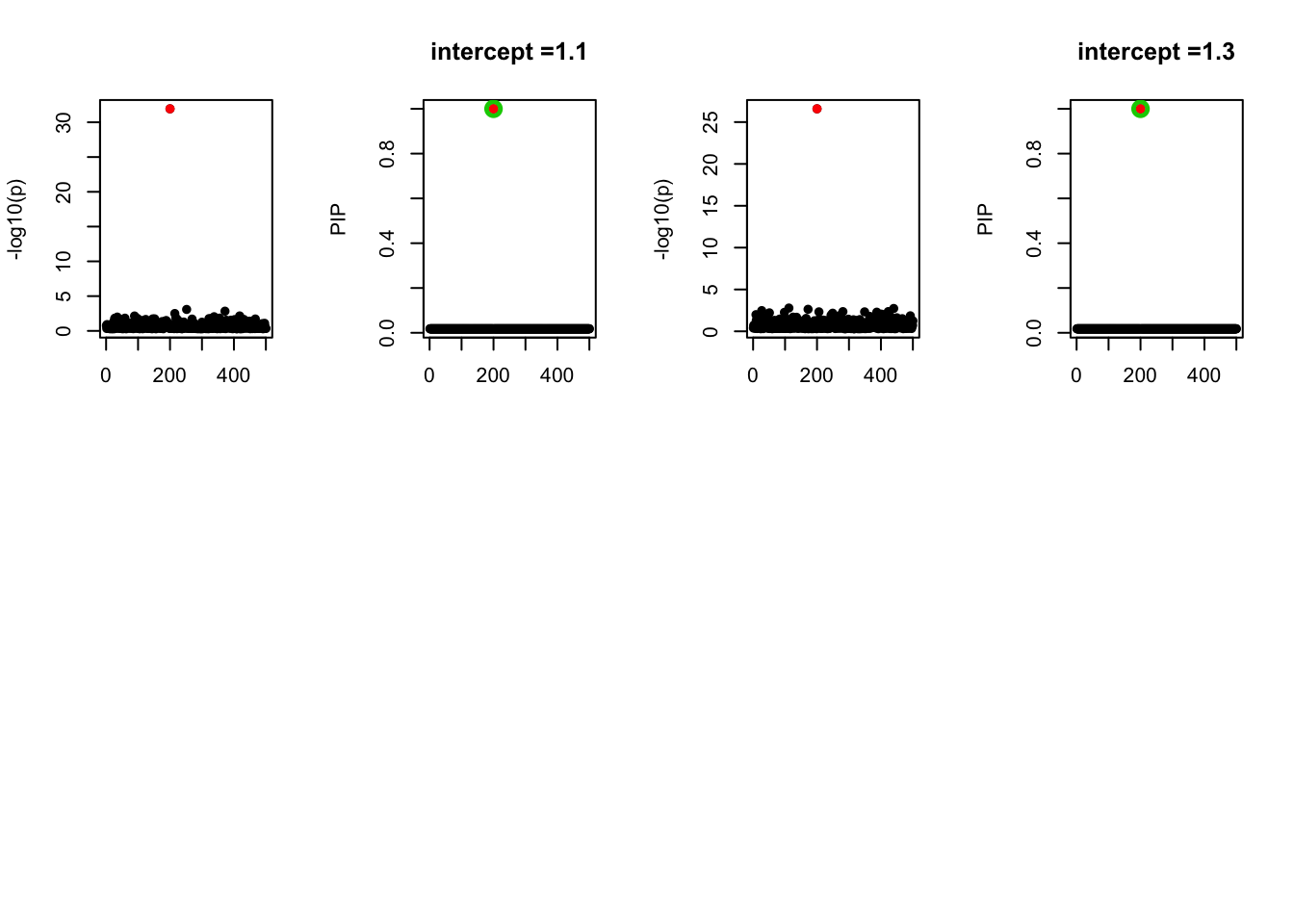
- Case 4: The number of true effects is 3 and the response y is simulated from the specified bernoulli model with intercept from -1.3 to 1.3.
par(mfrow=c(2, 4))
alpha = seq(-1.3,1.3,by=0.2)
set.seed(1)
for(a in alpha){
Y = rbinom(n, 1, exp(a+X %*% beta_true) / (1 + exp(a+X %*% beta_true)))
z = numeric(p)
for(i in 1:p){
z[i] = summary(glm(Y~X[,i], family = 'binomial'))$coef[2,3]
}
susie_plot(z, y='z', b=beta_true)
fit_z = susieR::susie_z(z, R, min_abs_corr = 0)
susie_plot(fit_z, y="PIP", b=beta_true, main=paste0('intercept =', a))
}

Expand here to see past versions of unnamed-chunk-10-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
Expand here to see past versions of unnamed-chunk-10-2.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
Expand here to see past versions of unnamed-chunk-10-3.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
Expand here to see past versions of unnamed-chunk-10-4.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
n \(<\) p
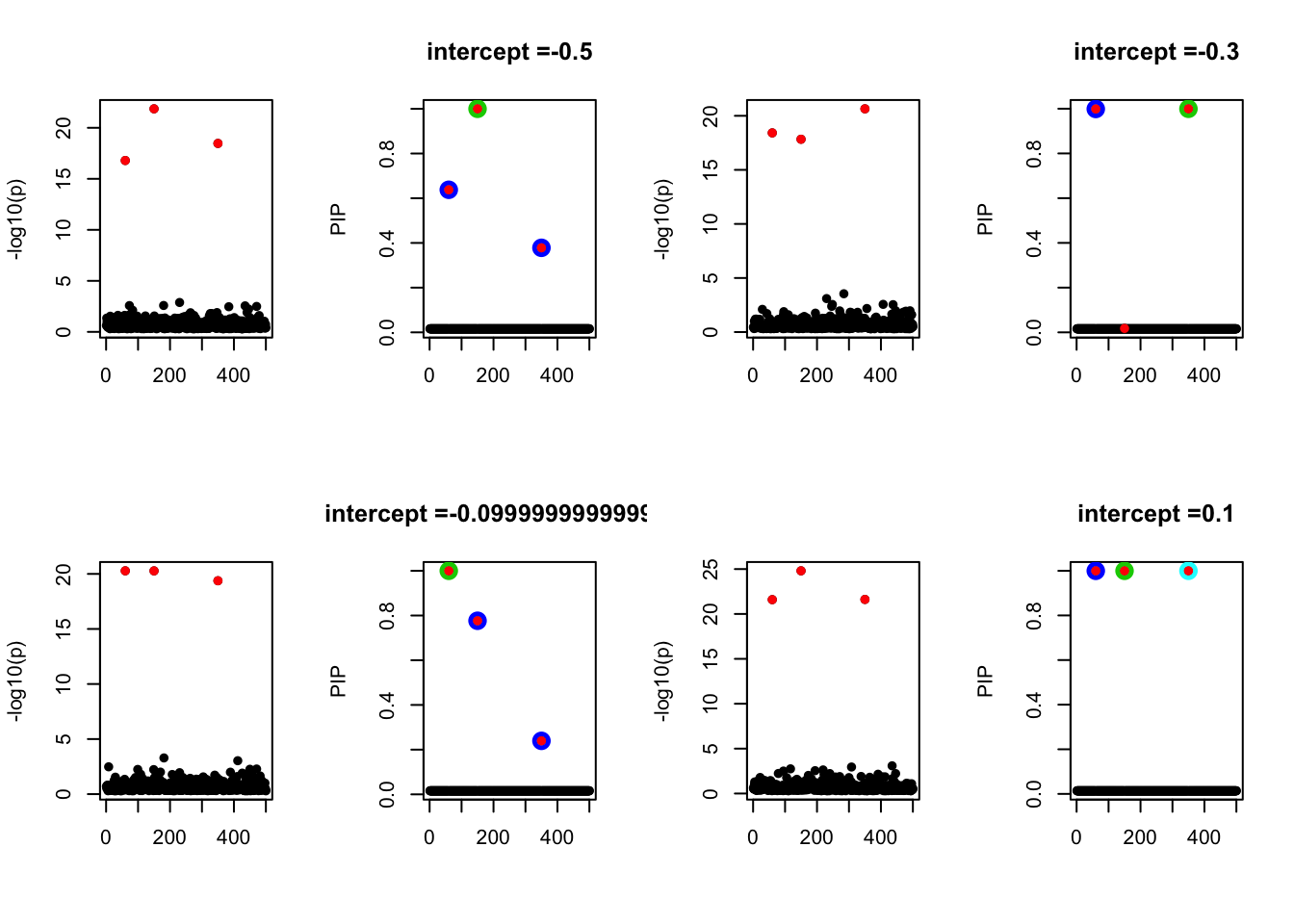
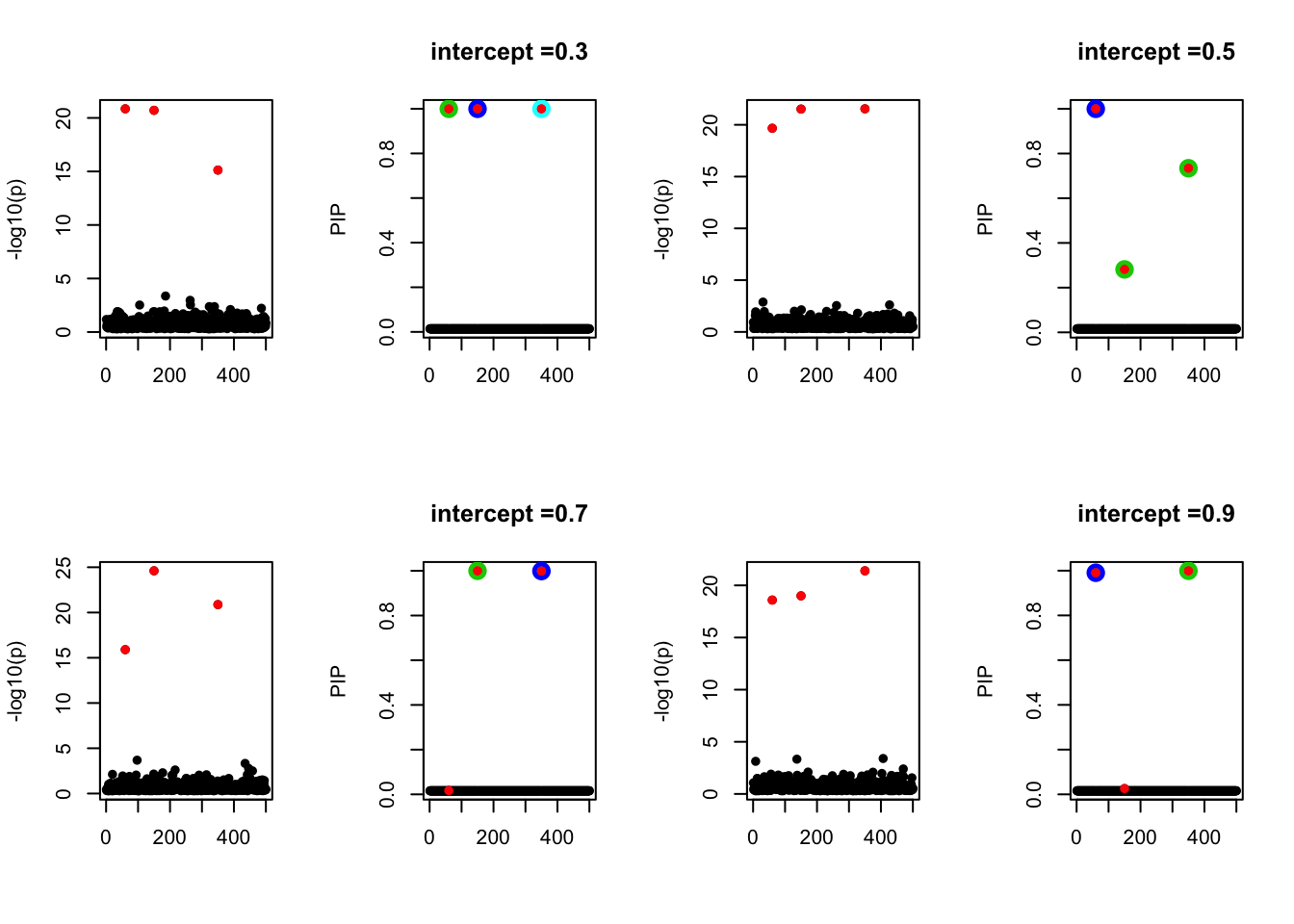
We run similations with n < p. Let n = 500, p=1000. The susie model does not capture the causal effects in all cases below.
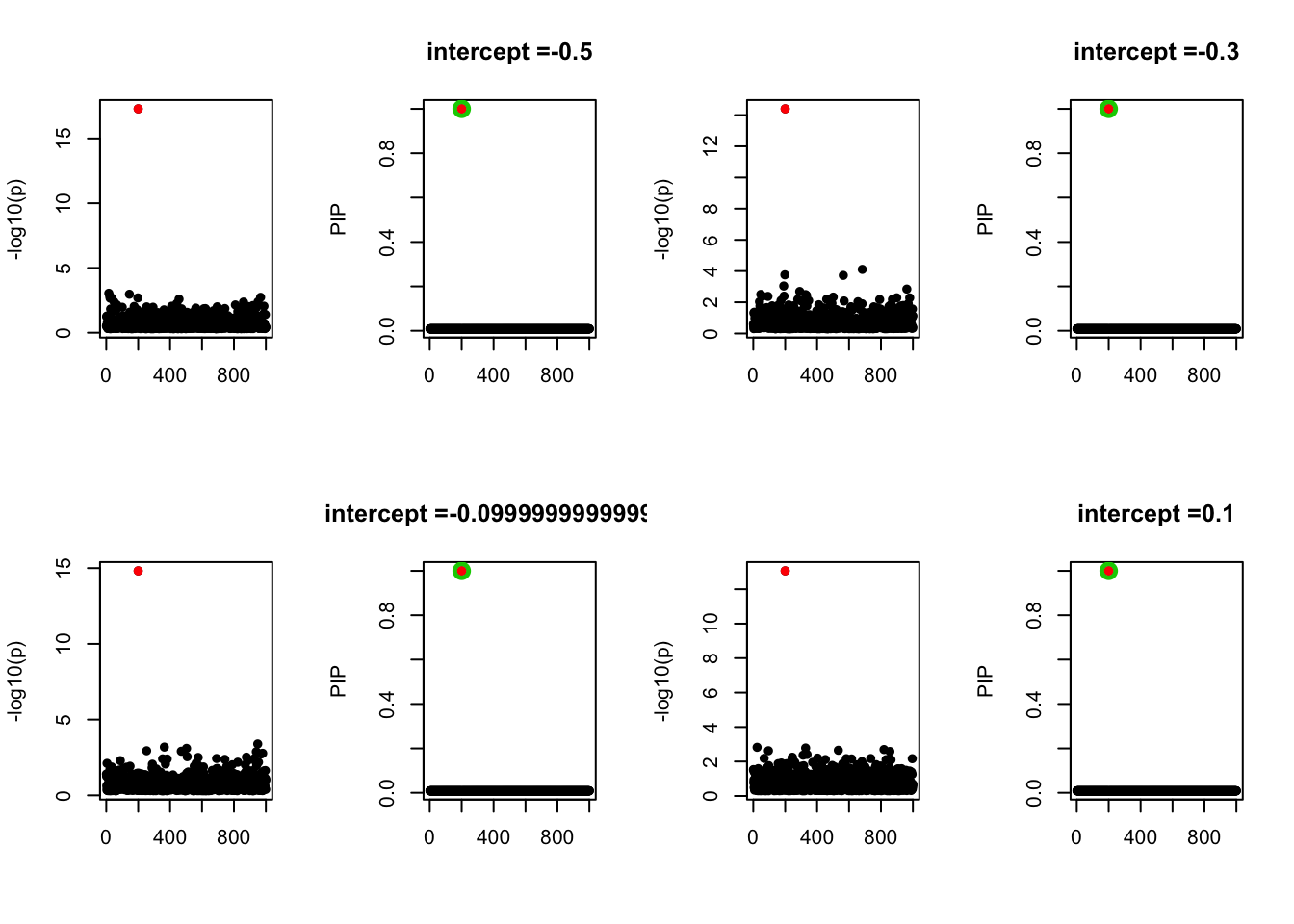
- Case 1: L=1. The true effect is b200. The response y is simulated from the specified bernoulli model without intercept.
set.seed(1)
n = 500
p = 1000
beta_true = rep(0, p)
beta_true[200] = 1
X = matrix(rnorm(n*p, 0, 1), nrow = n, ncol = p)
R = cor(X)
Y = rbinom(n, 1, exp(X %*% beta_true) / (1 + exp(X %*% beta_true)))
z = numeric(p)
for(i in 1:p){
z[i] = summary(glm(Y~X[,i], family = 'binomial'))$coef[2,3]
}
susie_plot(z, y='z', b=beta_true)

Expand here to see past versions of unnamed-chunk-12-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
|
cb3f4e5
|
zouyuxin
|
2018-12-05
|
fit_z = susieR::susie_z(z, R, min_abs_corr = 0)
susie_plot(fit_z, y="PIP", b=beta_true)

Expand here to see past versions of unnamed-chunk-14-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
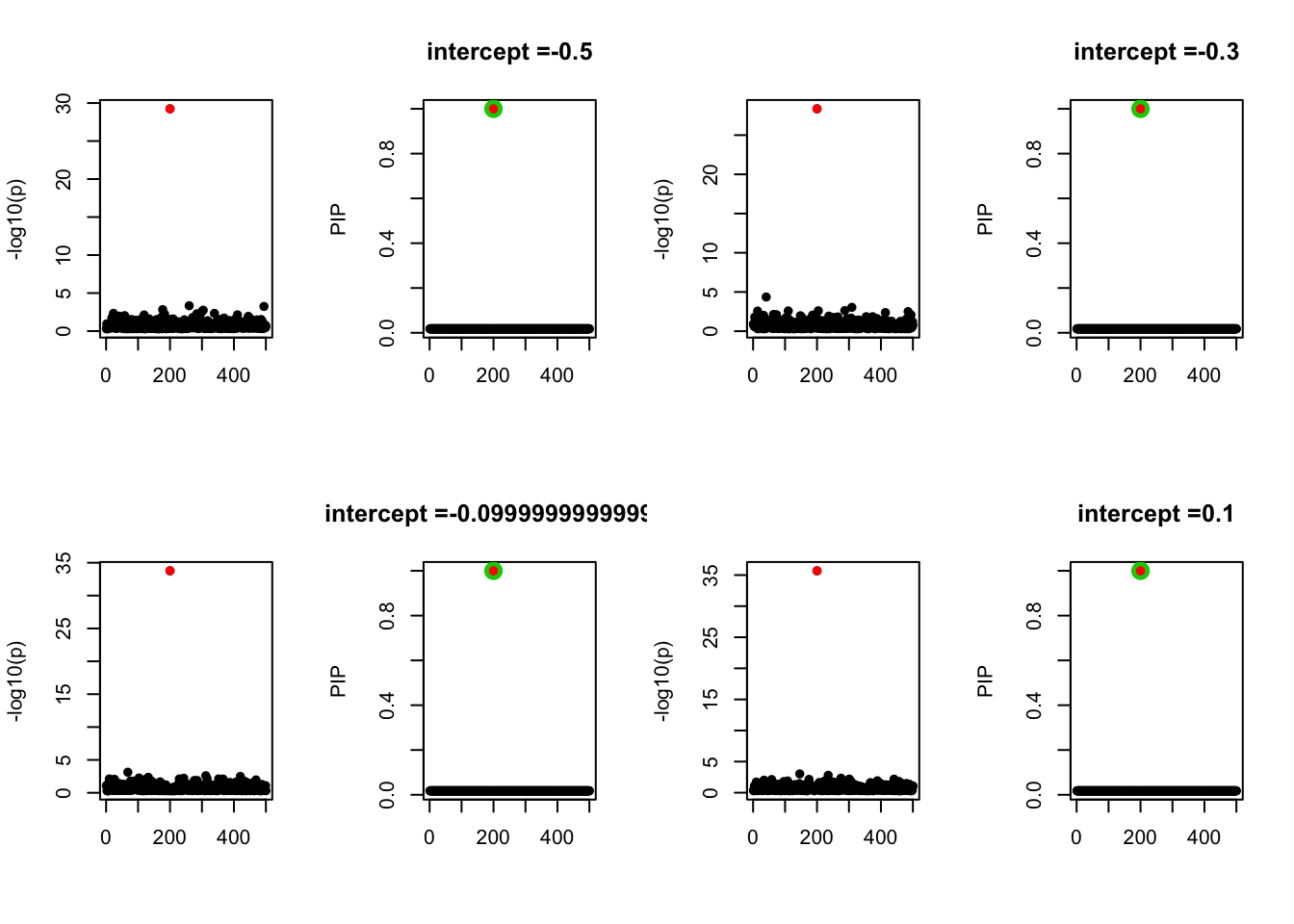
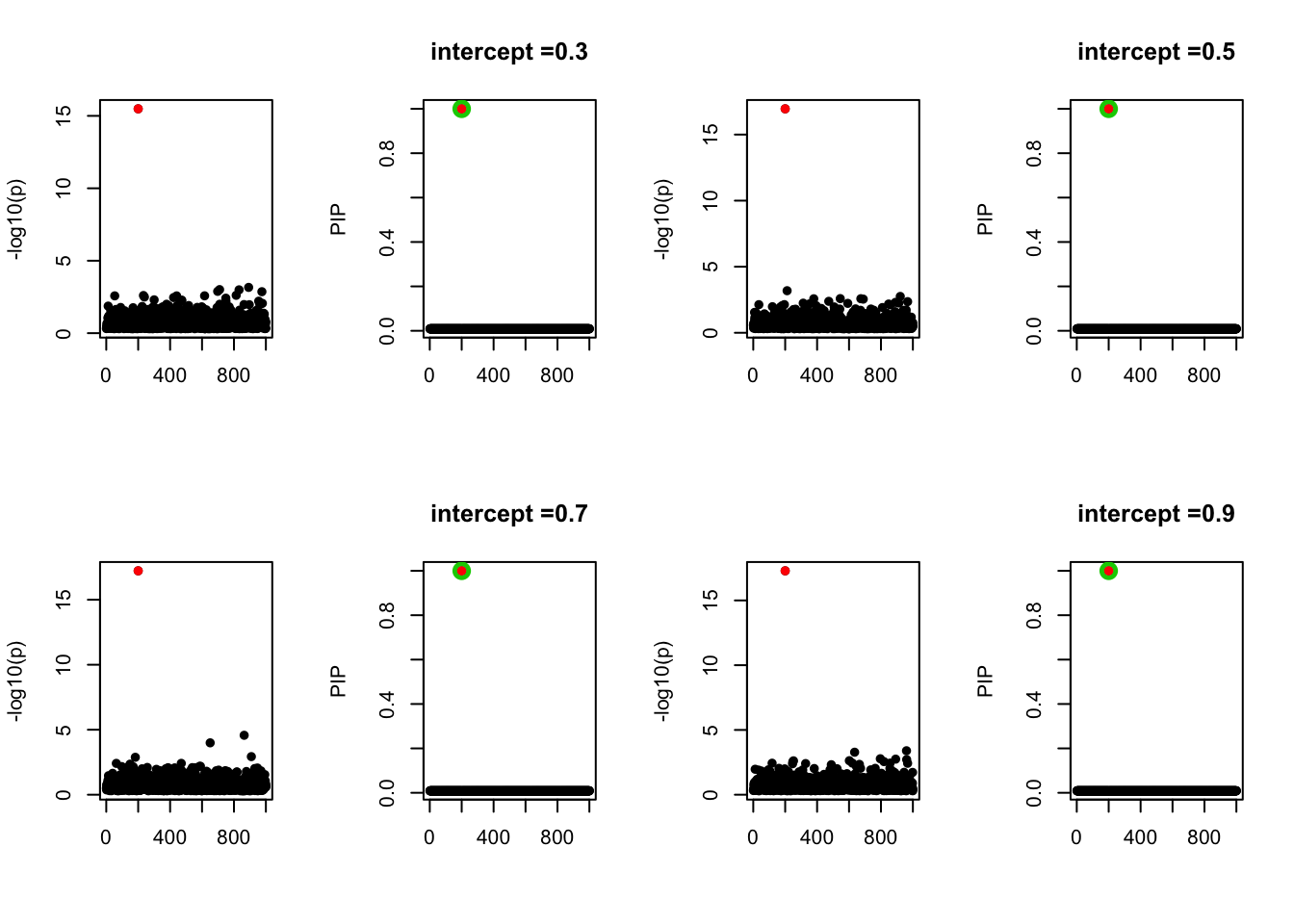
- Case 2: The response y is simulated from the specified bernoulli model with intercept from -1.3 to 1.3.
par(mfrow=c(2, 4))
alpha = seq(-1.3,1.3,by=0.2)
set.seed(1)
for(a in alpha){
Y = rbinom(n, 1, exp(a+X %*% beta_true) / (1 + exp(a+X %*% beta_true)))
z = numeric(p)
for(i in 1:p){
z[i] = summary(glm(Y~X[,i], family = 'binomial'))$coef[2,3]
}
susie_plot(z, y='z', b=beta_true)
fit_z = susieR::susie_z(z, R, min_abs_corr = 0)
susie_plot(fit_z, y="PIP", b=beta_true, main=paste0('intercept =', a))
}

Expand here to see past versions of unnamed-chunk-15-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
|
cb3f4e5
|
zouyuxin
|
2018-12-05
|
Expand here to see past versions of unnamed-chunk-15-2.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
Expand here to see past versions of unnamed-chunk-15-3.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
Expand here to see past versions of unnamed-chunk-15-4.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
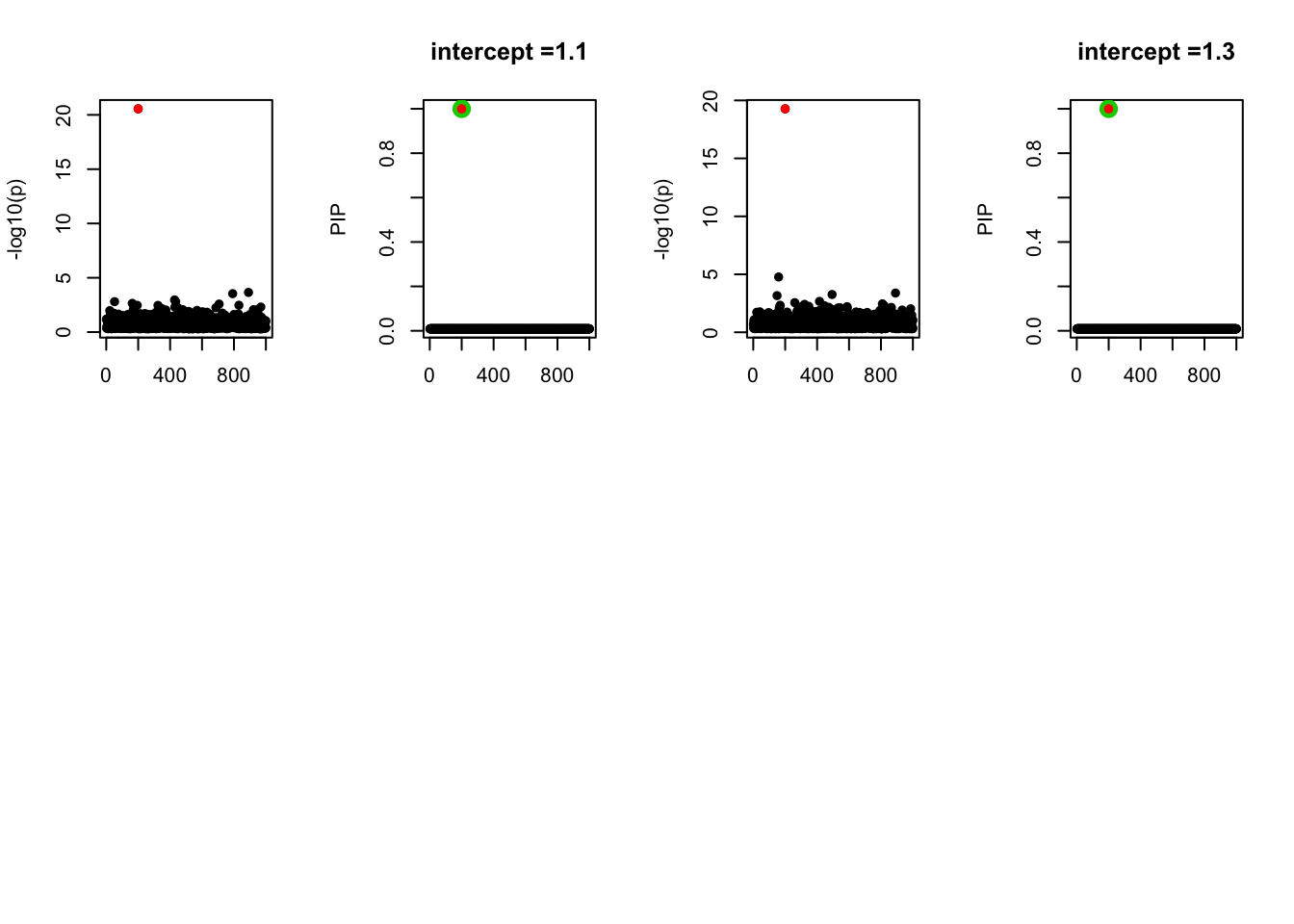
- Case 3: The number of true effects is 3. The true effects are b20, b100, b600. The response y is simulated from the specified bernoulli model without intercept.
beta_true = rep(0, p)
beta_true[c(20,600)] = 1
beta_true[100] = -1
set.seed(1)
Y = rbinom(n, 1, exp(X %*% beta_true) / (1 + exp(X %*% beta_true)))
z = numeric(p)
for(i in 1:p){
z[i] = summary(glm(Y~X[,i], family = 'binomial'))$coef[2,3]
}
susie_plot(z, y='z', b=beta_true)

Expand here to see past versions of unnamed-chunk-16-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
|
cb3f4e5
|
zouyuxin
|
2018-12-05
|
fit_z = susieR::susie_z(z, R, min_abs_corr = 0)
susie_plot(fit_z, y="PIP", b=beta_true)

Expand here to see past versions of unnamed-chunk-17-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
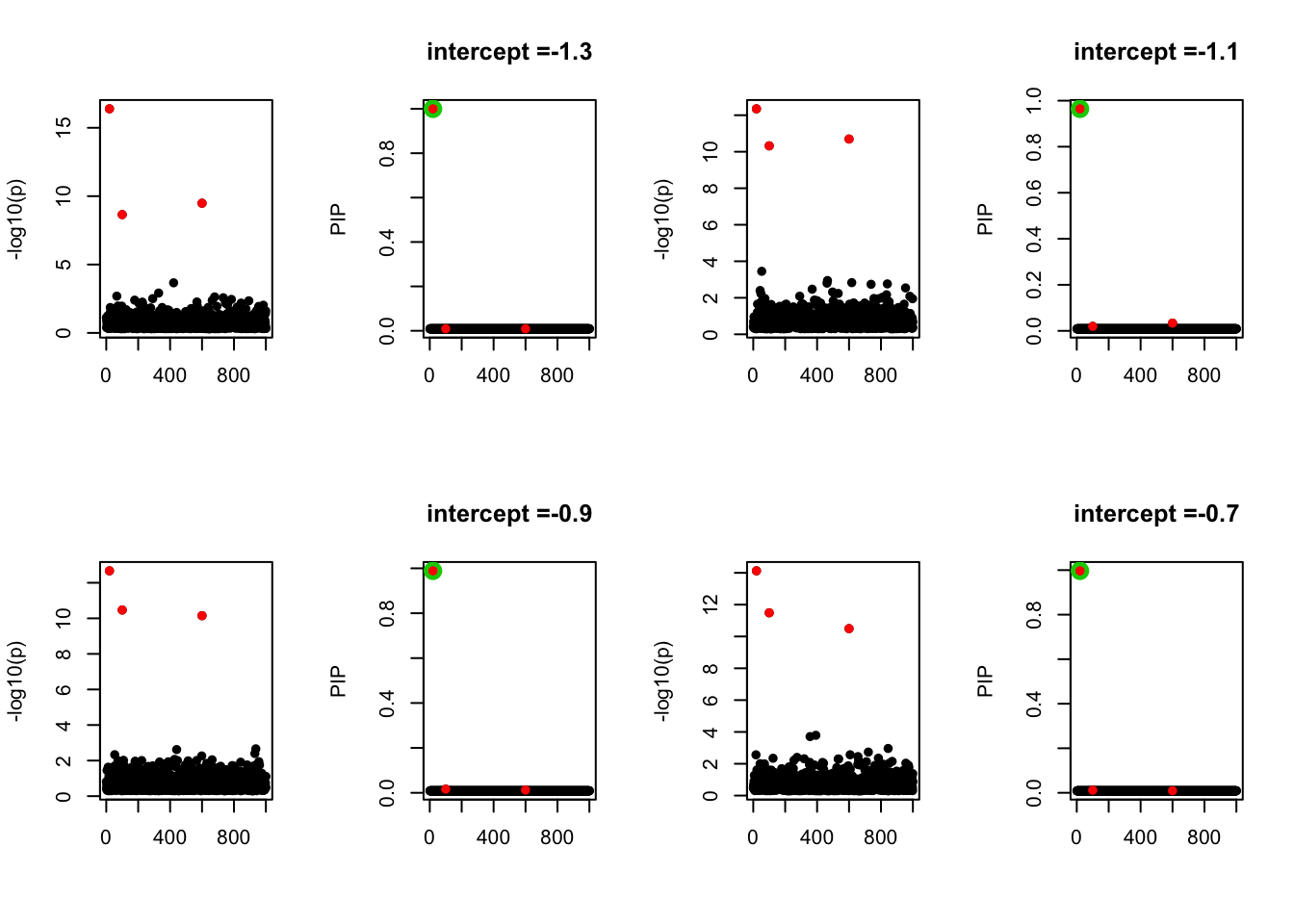
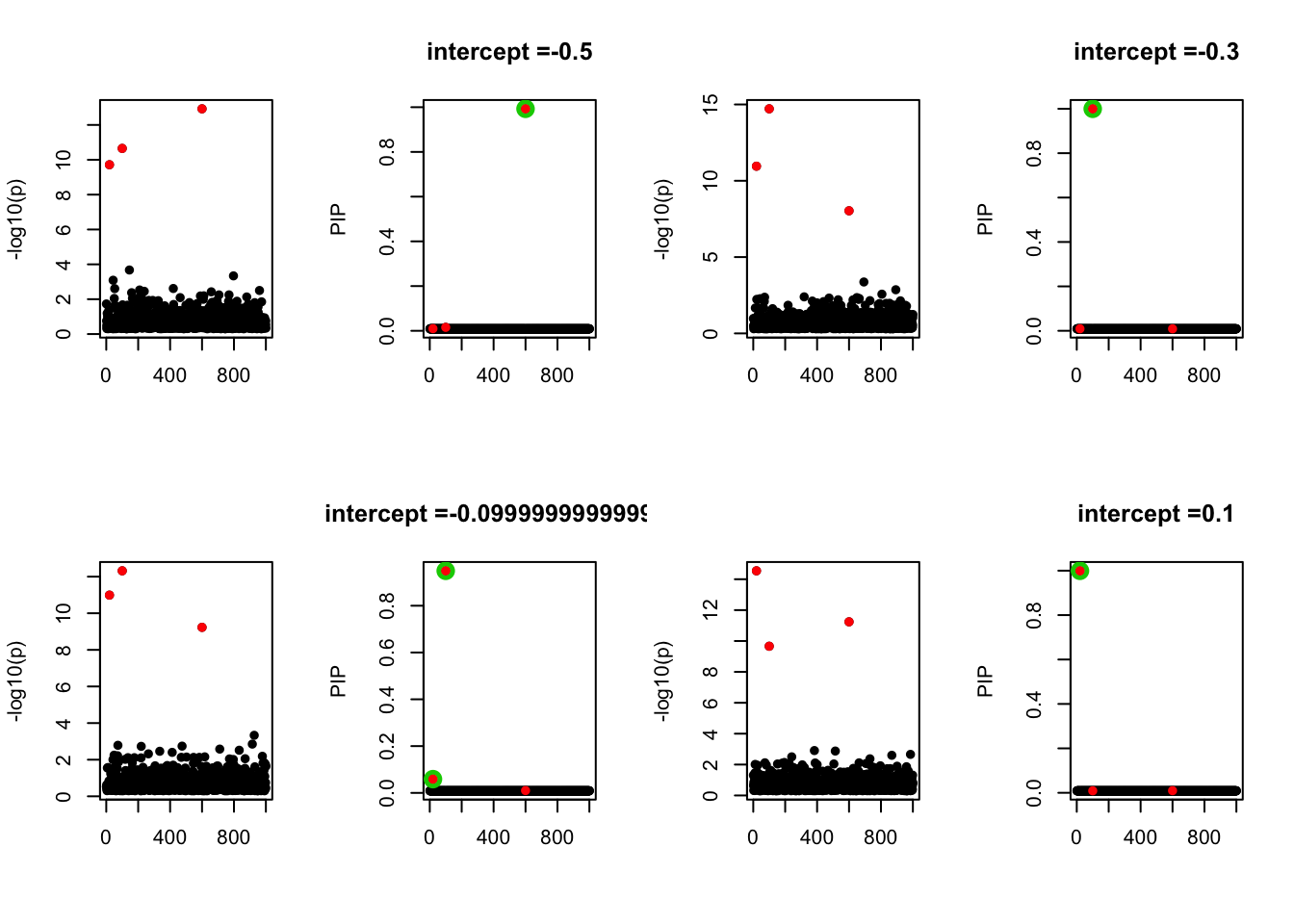
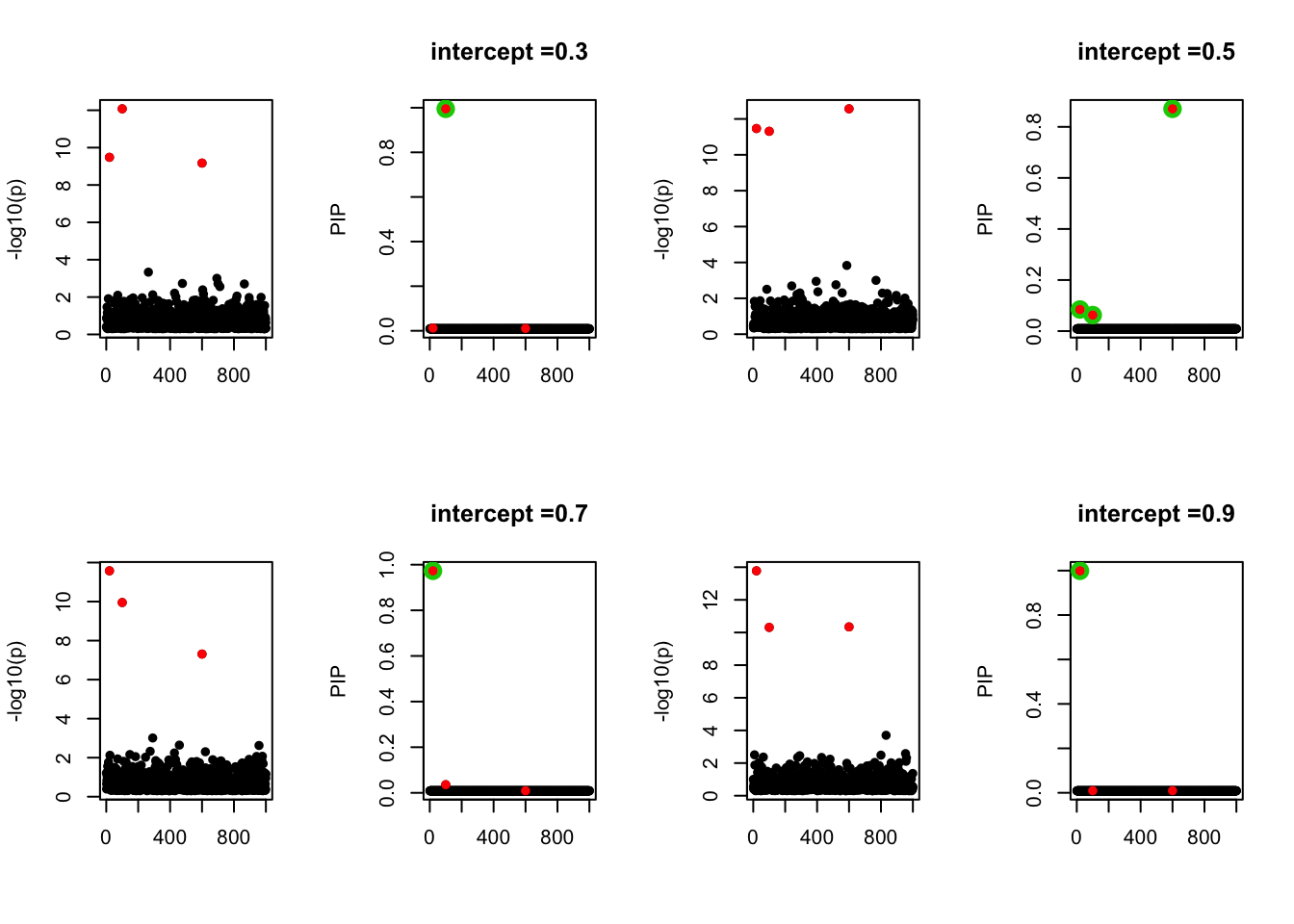
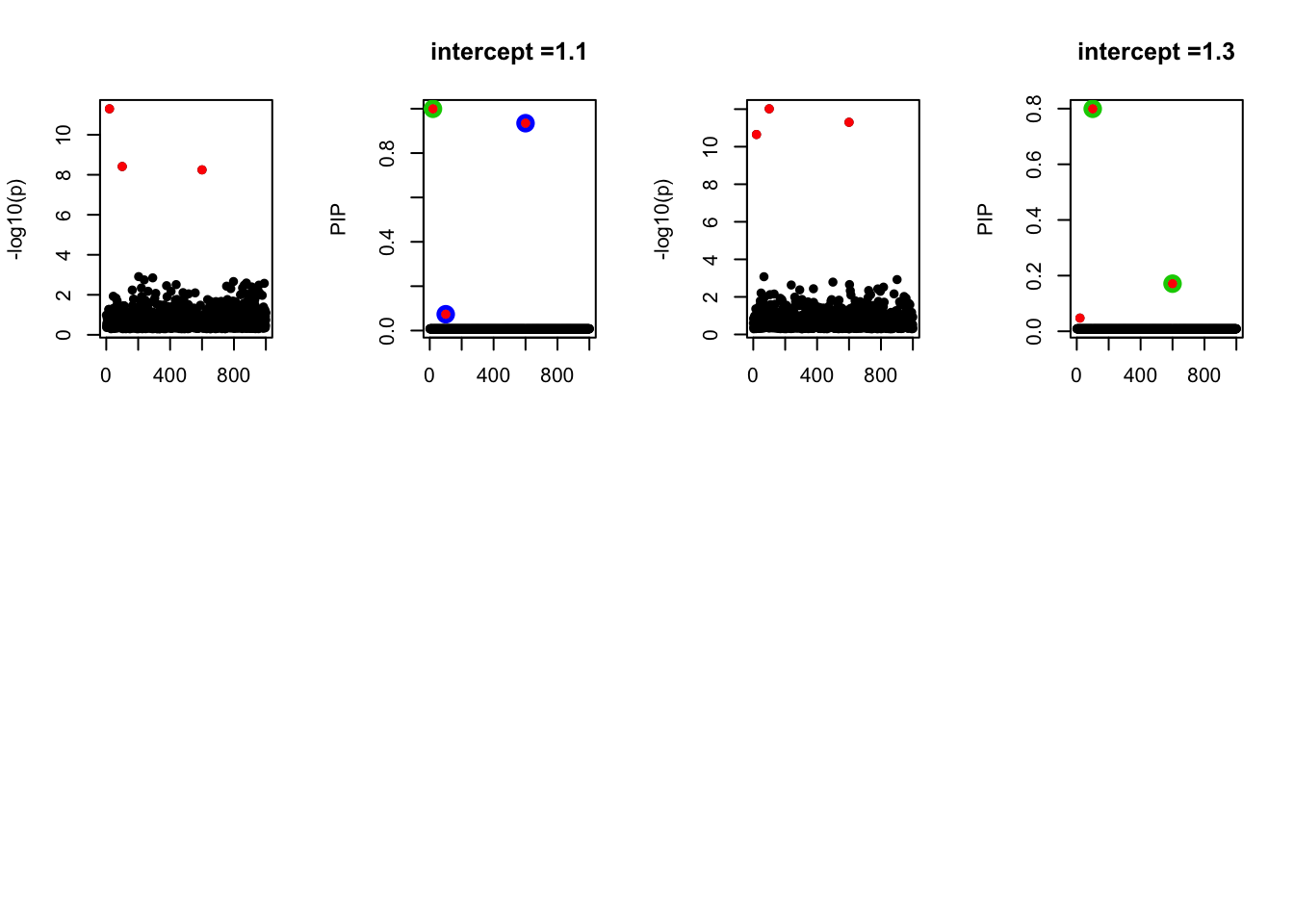
- Case 4: The number of true effects is 3 and the response y is simulated from the specified bernoulli model with intercept from -1.3 to 1.3.
par(mfrow=c(2, 4))
alpha = seq(-1.3,1.3,by=0.2)
set.seed(1)
for(a in alpha){
Y = rbinom(n, 1, exp(a+X %*% beta_true) / (1 + exp(a+X %*% beta_true)))
z = numeric(p)
for(i in 1:p){
z[i] = summary(glm(Y~X[,i], family = 'binomial'))$coef[2,3]
}
susie_plot(z, y='z', b=beta_true)
fit_z = susieR::susie_z(z, R, min_abs_corr = 0)
susie_plot(fit_z, y="PIP", b=beta_true, main=paste0('intercept =', a))
}
Expand here to see past versions of unnamed-chunk-18-1.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
|
cb3f4e5
|
zouyuxin
|
2018-12-05
|
Expand here to see past versions of unnamed-chunk-18-2.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
Expand here to see past versions of unnamed-chunk-18-3.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|
Expand here to see past versions of unnamed-chunk-18-4.png:
|
Version
|
Author
|
Date
|
|
003188b
|
zouyuxin
|
2018-12-10
|